Abstract
Predatory bacteria, like the model endoperiplasmic bacterium Bdellovibrio bacteriovorus, show several adaptations relevant to their requirements for locating, entering and killing other bacteria. The mechanisms underlying prey recognition and handling remain obscure. Here we use complementary genetic, microscopic and structural methods to address this deficit. During invasion, the B. bacteriovorus protein CpoB concentrates into a vesicular compartment that is deposited into the prey periplasm. Proteomic and structural analyses of vesicle contents reveal several fibre-like proteins, which we name the mosaic adhesive trimer (MAT) superfamily, and show localization on the predator surface before prey encounter. These dynamic proteins indicate a variety of binding capabilities, and we confirm that one MAT member shows specificity for surface glycans from a particular prey. Our study shows that the B. bacteriovorus MAT protein repertoire enables a broad means for the recognition and handling of diverse prey epitopes encountered during bacterial predation and invasion.
Similar content being viewed by others
Main
Bdellovibrio bacteriovorus is a bacterium that preys upon a wide range of other Gram-negative bacteria, entering their periplasm and consuming them from within. Physical contacts between B. bacteriovorus and diverse prey bacterial surfaces precede a decision-making stage in which the predator commits to invasion1,2,3,4. The invasion process requires interaction with, and traversal through, layers of prey lipopolysaccharide and peptidoglycan, requiring several different, transient, prey-adhesive steps. Contact regions between B. bacteriovorus and prey are apparent in both electron microscopy5 and tomography studies6, and proteinaceous adhesins would need to extend beyond predator lipopolysaccharide and be diverse enough to correspond to the breadth of molecular envelope diversity in known prey strains. The recognition of bacterial surfaces by viruses and the immune system is well documented, but the means by which bacterial predators recognize prey and engage in envelope traversal without causing lysis has remained obscure until now.
We found serendipitously that B. bacteriovorus Bd0635–mCherry (a homologue of CpoB, a PBP1b regulator in non-predatory bacteria7) moves dynamically inside invading predators, concentrating to a focus that develops into a discrete vesicular compartment that is deposited by the predator into the periphery of the prey periplasm, and persists in debris after predator exit. Analysis of vesicle contents reveals a family of phage-tail-fibre-like proteins with diverse adhesin domains.
These fibre-like proteins form discrete foci on attack-phase predators when invading prey, disappearing from external detection after predator invasion into prey, moving to a focus inside the prey periphery and co-locating with CpoBBd0635 for several family members. Structural studies reveal a variety of domain usages (and therefore binding capabilities) at the fibre C-termini, placing these on intertwined trimeric β-prisms. We infer that this large mosaic adhesive trimer (MAT) protein repertoire allows predator interactions with, and invasion of, a diversity of bacterial prey with different cell envelopes and surfaces. We confirm binding of one MAT type to multiple bacterial surface glycans including those of Proteus mirabilis.
Results
B. bacteriovorus deposits a predator-derived vesicle inside prey
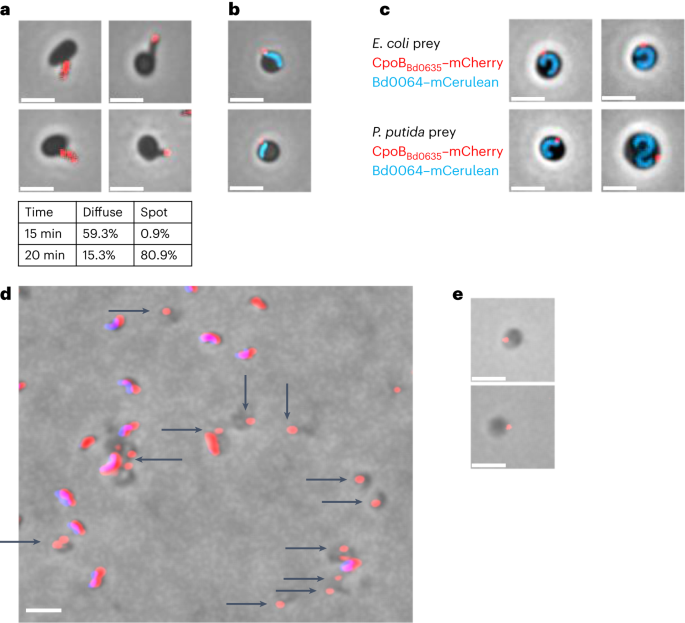
C-terminally mCherry-tagged CpoBBd0635 (Supplementary Fig. 1) was initially diffuse throughout predator cells but became focused near the flagellar pole of B. bacteriovorus as they invaded Escherichia coli prey (Fig. 1a and Supplementary Figs. 2–4). The mCherry focus was proximal to the trailing predator pole within the rounded prey ‘bdelloplast’ during early predation (Fig. 1b) but was detached from predator cells after 120–180 min of predator growth. At this later stage, the CpoBBd0635–mCherry-containing vesicle could be seen still within the periphery of bdelloplasts, but more clearly distinct from the B. bacteriovorus growing filamentous cell (Fig. 1c). It persisted robustly after prey exit, remaining fluorescent and attached to broken pieces of prey envelope debris, for wild-type predators (Fig. 1d), or persisting in prey cell ghost envelopes remaining after exit by mutant Δbd0468Δbd3279 double deacetylase predators (Fig. 1e)8. Deletion tests for bd0635 (refs. 2,9) resulted only in wild-type revertants, even when selecting for host-independent isolates, suggesting that CpoBBd0635 may be essential for viability in B. bacteriovorus.
a, Phase and epifluorescence microscopy tracking the movement of CpoBBd0635–mCherry during B. bacteriovorus attachment to, and entry into, prey. At earlier time points, most cells exhibit a diffuse fluorescent signal throughout the cell. The table shows that the fluorescence moves to become a distinct focus at the distal, flagellated pole of B. bacteriovorus as they begin to enter the prey cell. b,c, Phase and epifluorescence microscope images showing a fluorescent CpoBBd0635–mCherry focus (red) at the periphery of the prey cell, proximal to the cytoplasmic mCerulean of the predator cell (blue) at 30 min (b) and distal to the cytoplasmic mCerulean of the predator cell (blue) at 120–180 min (c) after the mixing of the predator and prey. This is shown when the prey is either E. coli or Pseudomonas putida. d, Phase-contrast and epifluorescence microscope images showing deposited vesicles remaining among prey envelope debris after prey exit: attack-phase B. bacteriovorus HD100 with CpoBBd0635–mCherry fluorescence (red) alongside DAPI predator genome fluorescence (blue—resulting in a pink merge) and broken prey bdelloplast debris lacking DAPI signal (dark grey), but in cases containing CpoBBd0635–mCherry fluorescent foci (black arrows). e, Phase and epifluorescence microscopy showing CpoBBd0635–mCherry fluorescent (red) foci left in the spherical ‘ghost’ prey bdelloplast peptidoglycan sphere (dark grey), which is specifically left behind, unbroken, after predation on E. coli by the mutant B. bacteriovorus Δbd0468Δbd3279. The images are representative of three independent repeats, and scale bars are 2 µm.
Vesicle CpoBBd0635–mCherry fluorescence and the HADA-labelled entry porthole in prey peptidoglycan1 were coincident (Supplementary Figs. 4–7). HADA, 3-[[(7-hydroxy-2-oxo-2H-1-benzopyran-3-yl)carbonyl]amino]-D-alanine hydrocholoride. Together, these results identify a robust vesicle that is produced by predators during prey entry and remains associated with the point of invasion in the bdelloplast.
Analysis of the vesicle reveals several phage-like proteins
Proteomic analysis of enriched CpoBBd0635–mCherry vesicle preparations (Supplementary Figs. 8 and 9, and ‘Methods’ in Supplementary Note 1) revealed a group of homologous proteins annotated as ‘cell wall anchor/YapH-like/phage-related tail fibre’ proteins (Supplementary Table 1). As discussed below, these could be grouped into two subfamilies, S74 and non-S74 (NS74), depending on the presence of an autoproteolytic S74 protease and chaperone domain10, with 13 and 8 members, respectively, identified in the B. bacteriovorus genome. For reasons explained later, we group these as the MAT family, for ‘mosaic adhesive trimeric’ fibre.
Several MAT proteins locate to the vesicle or bdelloplast
From this MAT grouping, we sampled several proteins for in vivo fluorescence tag localization. Bd2133 and Bd2439 (from the vesicle enrichment proteome) were deposited within the same vesicle as CpoB, while Bd3182 was deposited elsewhere within the bdelloplast and only at later time points (Fig. 2a; Supplementary Figs. 11, 12 and 15; and Supplementary Note 2).
a, Composite phase and epifluorescence microscope images showing attachment to, and growth of, predator strains inside E. coli prey bdelloplasts. The epifluorescence signal is from the C-terminally fluorescently tagged proteins: Bd1334–mCherry with CpoBBd0635–mTeal, Bd2133–mNeonGreen with CpoBBd0635–mCherry, Bd2439–mCherry with CpoBBd0635–mTeal, Bd2734–mTeal with CpoBBd0635–mCherry, Bd2740–mCherry with CpoBBd0635–mTeal and Bd3182–mNeonGreen with CpoBBd0635–mCherry. For comparison purposes, false colour is used in all images: CpoBBd0635 is coloured cyan and all MAT proteins are coloured red; coincident double fluorescence is shown in white. Representative images from a full predatory time course for each strain are presented in Supplementary Fig. 11. For Bd2133, Bd2439 and Bd2740, coincidence with C-terminally fluorescently tagged CpoBBd0635 is observed as a white focus (vesicle) indicated by yellow arrows. T15, T30 and T180 are 15 min, 30 min and 180 min, respectively, that have elapsed since predators and prey were mixed. Scale bars are 2 µm. The images are representative of three biological repeats. b, Composite phase and epifluorescence images showing spots of immunofluorescence on the outer surface of B. bacteriovorus attack-phase cells using FITC anti-mCherry antiserum and C-terminally mCherry-tagged MAT proteins. Bd2133 is absent from this analysis as a C-terminal mCherry tag was not tolerated by this protein. Details of immunofluorescent spot numbers and locations at different stages of predation are presented in Supplementary Fig. 13. The percentage of cells with observable immunofluorescent spots is presented for each strain below the image. Scale bars are 2 µm. The images are representative of cells from at least three biological repeats. c, Composite epifluorescence microscope images of FITC-labelled anti-mCherry antiserum immunofluorescence combined with the red membrane stain FM46-4 on attack-phase B. bacteriovorus cells reveal the locations of immunofluorescent spots with respect to the flagellum across all the mCherry-tagged MAT proteins examined. Details of the frequency of location for the different proteins are presented in Supplementary Fig. 13b. Scale bars are 2 µm. The images are representative of three biological repeats.
MAT NS74 proteins Bd1334, Bd2734 and Bd2740 were studied as they represented different subgroupings, with potentially different functionalities (discussed later), but had not been found in the proteome of the vesicle (which was collected after predation). We found that Bd2740–mCherry did locate to the CpoBBd0635-containing vesicle, deposited during predatory invasion, while Bd1334–mCherry and Bd2734–mCherry fluorescence were diffuse throughout the B. bacteriovorus cell (Fig. 2a, and Supplementary Figs. 11 and 12).
MAT proteins are surface displayed on predators
We hypothesized that some MAT proteins may present their C-termini out of the B. bacteriovorus cell for prey interactions. Testing with a fluorescent secondary antibody and anti-mCherry antiserum revealed that all C-terminally tagged MAT proteins tested were detected at various locations on the surface of the attack-phase B. bacteriovorus cell, as expected for prey-interacting fibres, but were not detected externally after predator entry into the bdelloplast (Fig. 2b,c, Supplementary Figs. 10 and 12–16, and Supplementary Note 2).
We deleted each individual gene for the six sampled proteins, finding a small, but significant, predation defect in two of the (NS74) mutants: Δbd2734 and Δbd2740, using a microtitre-plate-based predation assay11 (Fig. 3). That the effect was small, and not seen for all MAT mutants (Supplementary Fig. 17), was expected as a multifactorial recognition and handling of the prey cell surface occurs during B. bacteriovorus predation. The rate of predation (rMax) on E. coli S17-1 prey was reduced with mutants Δbd2734 and Δbd2740, but predation on an alternative prey, Proteus mirabilis, was not significantly different for any MAT mutant compared with the wild type (Fig. 3b). The reduced rMax may be a result of delayed attachment to, and entry into, prey; this is supported by our observation that detectibly fewer prey had been entered by the Δbd2734 (90.5% ± −4%) and Δbd2740 (91.1% ± −1.6%) mutants 30 min after mixing compared with the wild type (WT for Δbd2734, 96.7% ± −0.7%, and WT for Δbd2740, 96.7% ± −1.4%; Fig. 3c).
a, Monitoring reductions in prey cell OD600 as a result of prey lysis by predator MAT mutants (red) relative to the wild type (blue) versus prey E. coli only without predators (Ec) (green). Examples shown are predation by Δbd2439, which has no predatory defect, and Δbd2734 and Δbd2740, which are delayed in predation relative to the wild type. b, Measures of the rMax for all mutant strains. rMax is significantly higher (a lower rate of OD600 reduction and therefore predation) for Δbd2734 and Δbd2740 when preying upon E. coli (Kruskal–Wallis test on non-normally distributed data, **P < 0.01; P = 0.0099 for Δbd2734 and P = 0.005 for Δbd2740), but not significantly different for any strains preying upon Proteus mirabilis (one-way ANOVA on normally distributed data). Values of n (independent biological repeats) are, for predation on E. coli, 4 (Δbd2133, Δbd2439), 5 (Δbd1334, Δbd2734, Δbd2740) and 6 (HD100, Δbd3182) and, for predation on Proteus mirabilis, 3 (Δbd3182) and 4 (all others). c, Percentage of bdelloplasts visually scored for a fully entered B. bacteriovorus 30 min after the mixing of the predator and prey for B. bacteriovorus HD100 wild type versus MAT deletion mutant strains Δbd2734 and Δbd2740. The P values presented are from two-tailed unpaired t-test. Data are from four biological repeats. Details in Supplementary Fig. 17b. d, Example regions of interest showing the diversity of cells characterized as fully entered and incomplete for B. bacteriovorus MAT mutant entry. Cells counted as incomplete entry included rod-shaped prey with B. bacteriovorus attached (i) and rounded prey with partially entered B. bacteriovorus (ii) as indicated by yellow arrows. Blue arrows indicate cells characterized as B. bacteriovorus having fully entered the periplasm of the prey. Scale bars are 2 µm.
Characterization of the wider MAT family and S74 activity
Having found MAT members on the surface of predators and some in the vesicle proteome, we sought to identify the full complement, and their structural diversity, in B. bacteriovorus HD100. Using Position Specific Iterative BLAST12, manual inspection and fold prediction13, we were able to find 21 confident matches (Supplementary Table 1), 13 terminating in a consensus S74 peptidase domain with a SDxxxK catalytic motif10 and 8 NS74 MATs that do not.
The S74 protein domain is a dual chaperone and protease domain shown to confer quality control, ensuring productive trimerization in surface-exposed phage β-helical spikes and enzymes14. All 21 MATs contain a conserved β-rich domain of ~130 aa immediately following the signal peptide.
A C-terminal region (amino acids 632–922) of Bd3182 confirmed the predicted chaperone and protease activity of the S74 domain, yielding the expected two bands, with self-cleavage at catalytic residue S782 (Fig. 4a). An S782A null mutant results in a single unprocessed species (Fig. 4a), in a manner identical to other S74-containing proteins10. Bd3182–mCherry prey bdelloplast location, sometimes as a tight focus, was seen at 120–180 min into predation (Supplementary Fig. 15) and was abolished in the S782A variant. Having potentially found a family of trimerizing adhesins with diverse adhesive tips facilitating predator–prey interactions, we set out to define their structures.
a, ColabFold model of Bd3182 β-helix and S74 chaperone. The accompanying SDS-PAGE gel shows classical S74 processing of Bd3182 into separate fibre and chaperone domains, which is blocked in the S782A mutant. This autoproteolysis was observed on more than four independently expressed samples of Bd3182. b, Crystal structure of Bd3182632–782 (hereafter referred to as Bd3182). The single chain (left) forms a symmetric trimer (right). Distinctive motifs form capping loops on the β-helix edges (yellow). c, The capping loops form similar structures exemplified by superposition of the proximal (green) and distal (blue) loops of Bd3182. An alignment of capping loops from multiple fibres (Supplementary Fig. 18) provides a consensus motif for this feature (bottom). d, The phage T5 L-shaped tail fibre (red; PDB code 4UW8 (ref. 14)) and K1F endosialidase (green; PDB code 3GW6 (ref. 10)) show similar β-helical structures to that of Bd3182. The surface of each features a cleft formed by the beta sheet of the fibre (orange surface). e, The surface of Bd3182 shows a continuous cleft (orange), capped by the proximal (green) and distal (blue) capping loops. Bacteriophage P22 (structural homologue of Bd3182; main text) binding of O-antigen (carbon atoms, yellow) within a cleft on the surface of the β-helical fibre (PDB code 3TH0 (ref. 50)). f, Full-length fibre of Bd3182 modelled using a composite of ColabFold models and our experimentally determined crystal structure.
Structure of a representative S74 group MAT fibre tip
We generated three different forms of the mature ‘tip’ region of family member Bd3182 (Fig. 4a and Supplementary Table 2; we describe the 1.1 Å form herein). The structure traces P647 to A781, forming a trimeric β-helix 40 Å across and 60 Å high (Fig. 4b). Each face of the β-helix is formed from multiple chains, resulting in an intertwined structure. Bd3182 retains the classical hydrophobic β-helix core, and the arrangement of loops on the surface creates a 30 Å × 15 Å cleft (Fig. 4b), limited in dimensions by two capping loops of identical structure (R656–H669 and F752–H765; Fig. 4b,c).
We found related loops in other B. bacteriovorus MATs (Supplementary Fig. 18) and generated a consensus motif (V–G–G/N–π–X–X–P–X–X–X–L; Fig. 4c). Searching the Protein Data Bank with this motif returns a hit to the S74-containing Myelin Regulatory Factor (MyRF)15, revealing an S74 feature conserved beyond the catalytic protease.
Trimer-to-trimer comparison reveals homology to bacteriophage S74 proteins, with Root-Mean-Square Deviations of 2.4 Å and 2.5 Å for phage K1F (3GW6 (ref. 10)) and bacteriophage T5 (4UW8 (ref. 14)), respectively. The RMSDs indicate that Bd3182 is a true S74 member, with a surface cleft similar to that of others (Fig. 4d,e), whose exposure (via S74 proteolysis) is necessary to bind their O-antigen ligand. The best match of a monomer via DALI16 analysis is to the podovirus P22 tailspike (RMSD 4.4 Å), whose bound O-antigen fragment17 is positioned in agreement with the Bd3182 cleft (Fig. 4e). An AlphaFold18 model of full-length Bd3182 reveals that trimerization is retained along its entirety, generating a fibre ~550 Å long (Fig. 4f and Supplementary Fig. 19). Thus, predatory bacterial MAT protein Bd3182 is related to phage adhesins but uses a distinctly longer fold with a more exposed binding cleft.
Structures of NS74 members reveal MAT fibre-tip diversity
Taking Bd3182 as a representative of the other S74-terminating MATs, we next studied the eight more divergent ‘NS74’ proteins with differing termini. A region comprising N662 to the C-terminus of Bd2133 was determined to 2.5 Å resolution (Supplementary Table 2).
The Bd2133 tip is 150 Å long, differentiated into three subdomains (Fig. 5a)—two β-helix regions akin to Bd3182 (G683–E813 and N814–A907) and a β-sandwich domain that substitutes for the S74 chaperone (W913–K1031). The latter two domains are linked by a stalk-like linear peptide (A906AGAAT). Despite a general agreement in the β-helix region, Bd2133 does not possess a Bd3182/S74-like cleft. Equivalents to the Bd3182 capping loops are present, demonstrative of common principles in the S74 and NS74 subgroupings.
a, Crystal structure of Bd2133662–1,031 (hereafter referred to as Bd2133). This portion of the trimeric fibre forms a structure 142 Å in length and can be subdivided into three distinct domains: two β-helix domains and a C-terminal β-rich tip domain. b, The β-rich tip domain of Bd2133 forms a previously uncharacterized fold comprising a β-sandwich that packs into a trimer with multiple atoms and ions along the central three-fold axis (left). A stick representation of the coordination is shown (right), with a 1.5 Å 2fo–fc map at 2σ. c, The N-terminus of Bd2133 (residues T681SGNLG; orange) binds to the C-terminal domain in the crystal structure (left). The crystal symmetry provides a 1:1 stoichiometry of the N-terminal peptide to the trimeric C-terminal domain. The peptide binds through multiple backbone and side-chain hydrogen bonds, and packs L685 into a moderately hydrophobic shallow pocket. d, Bd213321–1,031 full-length fibres show flexibility when imaged via negative-stain electron microscopy. These are representative micrographs from multiple grids made of a single sample of purified Bd2133. e, The crystal structure of Bd1334818–1,151 (hereafter referred to as Bd1334). This portion of the trimeric fibre forms a structure 95 Å in length, and the protein can be subdivided into a partial β-helix domain, a full β-helix domain and a C-terminal β-rich tip domain. f, The Bd1334 β-rich tip domain forms a previously uncharacterized fold comprising a squashed β-sandwich that packs into a trimer with the strands diagonal to the trimeric axis. A single disulphide is present (right, shown as spheres). g, A phosphate molecule (shown as spheres) is buried in a cavity formed between two protomers. Multiple residues interact with the phosphate, including W1078 and W1102, which form a cage around the ion. h, Full-length fibres of Bd2133 and Bd1334 modelled using a composite of ColabFold13 models and our experimentally determined crystal structures. The two proteins comprise very similar N-termini, both in sequence (75% sequence ID) and predicted structure; however, the C-terminus has been exchanged at some point during evolution.
The β-sandwich tip domain of Bd2133 adopts a twisted fold, trimerizing about strand G989–W999 (burying 5,640 Å2 per monomer). There are several atoms lying on the three-fold axis (Fig. 5b). Solving a 1 Å tip-only variant (A910 onwards) allowed us to confidently model these as a water, a sodium and a chloride ion. Unexpectedly, there are no homologues of this domain in the PDB (judged by DALI16), but FoldSeek19 found strong homology to Synechococcaceae and Competibacter proteins (Supplementary Fig. 20). During Bd2133 refinement, we noticed additional density at the sandwich tip, where the trimer interface creates a pocket using Y985, N956 and N1020. Guided by crystal symmetry, we have modelled this as an N-terminal hexapeptide derived from a nearby copy (Fig. 5c).
Negative-stain electron microscopy of full-length Bd2133 (residues 21–1,031) confirmed fibre formation, with dimensions commensurate with an AlphaFold18 model (Fig. 5d,h) and our partial structural determination, showing fibre conformational flexibility (Fig. 5d).
Bd1334 shows chimeric MAT-tip architecture
To address the potential for domain-based diversification, we crystallized the tip of Bd1334 (which shares 75% identity with Bd2133 in the fibre region but then diversifies), from S818 and S914 onwards, diffracting to 2.6 Å and 1.4 Å resolution, respectively (Supplementary Table 2). The structure is composed of three subdomains (Fig. 5e–g): a partial β-helix (D823–A868), a full β-helix (T869–Q1011) and a flat β-rich tip (P1012–W1151). The tip of Bd1334 has no similarity to that of Bd2133, forming a β-sandwich of a completely different, square-like shape. Tip trimers are mediated by edge-to-edge contacts, burying 6,900 Å2 per monomer. A composite pocket is formed from two chains, making a tryptophan cage with W1078 and W1102 from one monomer and the terminal W1151 from the opposing monomer (Fig. 5g). W1102 has an unusual cis peptide bond often enriched in binding sites20. There are no structural homologues for Bd1334 in the PDB, and sequence homologues are limited, with sequence conservation patterns being non-informative. Thus, the Bd1334 data show that MAT tips can be swapped out in evolution (Fig. 5h) and that Bd1334 is probably specific for binding an epitope encountered and bound only by B. bacteriovorus bacteria.
Extension of tip diversification into known adhesive folds
We next characterized two more divergent NS74s, Bd2439 and Bd2734, obtaining a tip-only variant of Bd2734 (residues T691–L843, 1.8 Å) and a longer construct of Bd2439 (G837–D1107, 1.6 Å; Supplementary Table 2). The resulting structures reveal that both proteins use a related fold (Fig. 6a,b).
a, The crystal structures of Bd2439837–1,107 and Bd2734691–843 (hereafter referred to as Bd2439 and Bd2734, respectively) reveal the presence of TNF-like domains at the C-terminus of both fibres. b, The two TNF-like domains are shown in the same orientation and coloured as a Jones rainbow to highlight the structural homology of the two domains (disulphides shown as spheres). c, The electrostatic potential of the Bd2439 TNF-like domain trimer surface shows a positively charged pocket between two protomers. d, GlcNAc-bound Bd2439 (left). Two tyrosines flank the sugar moiety, which orientates its C3 and C4 hydroxyls into the charged pocket to hydrogen bond with two His and two Arg residues (right). e, Glycan array screening of Bd2439 against various bacterial LPS (lipopolysaccharide). The bars show the mean of four experiments ± s.d. Only assays in which binding was statistically significant against the negative control are shown (**P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001 by one-way ANOVA with Dunnett’s test using GraphPad Prism 8.0; n = 4 technical replicates). Binding was significant for E. coli K235 (P < 0.0001), E. coli EH100 (Ra mutant), Salmonella enterica typhimurium (P = 0.006), Serratia marcescens (P < 0.0001) and Porphyromonas gingivalis (P = 0.0089), and particularly strong binding was observed to Proteus mirabilis LPS (P < 0.0001). f, The crystal structure of Bd2740518–627 (hereafter referred to as Bd2740), which uses a trimeric C-type lectin fold. g, Two protomers of Bd2740 are shown (red and white) superposed with the structurally homologous snake venom lectin agkisacucetin (PDB code 3UBU51). The fold shows good structural alignment with an RMSD of 2.48 Å over 96 residues, with the major difference being the shift of helix B, which is used by Bd2740 for the trimer interface. h, Comparison of Bd2740 (left) with IX-BP (right; the highest-similarity Ca2+-binding homologue; PDB code 1BJ3 (ref. 30)) shows that Bd2740 does not bind calcium owing to the alteration of helix B and the lack of calcium-binding residues in this region. i, Full-length fibres of Bd2439, Bd2734 and Bd2740 modelled using a composite of ColabFold13 models and the experimentally determined crystal structures. These models highlight the different lengths of the fibres, and both Bd2734 and Bd2740 lack the β-helical domains of the other fibres.
Bd2439 is modular, formed by a β-helix N850–V968, a R969 linker that coordinates the carboxy terminus and a compact β-domain tip from S970–D1107 (Fig. 6a). Structure searches with DALI16, FoldSeek21 and oligomer-sensitive TopSearch22 all confirm similarity to trimeric members of the tumour necrosis factor (TNF) family (for example, RMSD 2.3 A for human BAFF, 6FXN23), typically receptors and lectins24. The Bd2734 tip represents a divergent TNF fold, with large pockets between monomers (Fig. 6a,b). The modified fold of Bd2734 affords it closer similarity to a different TNF subgroup, with the best match to a fucose-specific lectin from Burkholderia (2WQ4 (ref. 25), 5% sequence identity, 2.3 Å RMSD) and a C1Q-like family protein (4QPY26, 15% identity, 2.7 Å RMSD). These two MAT tips are therefore more similar to other TNF proteins than they are to one another (RMSD 3.1 Å). Superimposing with a Burkholderia lectin–fucose complex25 shows how the Bd2734 pocket can accommodate a terminal monosaccharide (Supplementary Fig. 21). Mapping a sequence alignment (Supplementary Fig. 22) to Bd2439 identifies a conserved interfacial pocket (Y1031, R1040, E1044, H1053 and H1055 from one monomer; R1022 and Y1097 from another; Fig. 6c). These residues coordinate two hydroxyl groups from a bound ethylene glycol (Supplementary Fig. 23). Testing a variety of saccharides, we observed binding of Bd2439 to GlcNAc (from a GlcNAc–MurNAc disaccharide; 1.84 Å resolution dataset), revealing that the above residue cluster recognizes two adjacent ring hydroxyls (Fig. 6d). Thermal unfolding experiments confirm this specificity, revealing a biphasic unfolding whose second phase is shifted in the presence of sugars that contain these features (Supplementary Fig. 24).
We next determined the structure of Bd2740 (residues 517–626, 2.6 Å resolution), whose fold was previously cryptic. Our resulting structure reveals a variant of the C-type lectin fold (Fig. 6f), a widely dispersed superfamily named after its use of calcium to bind sugars, proteins and lipids27. The fold has many structural homologues (Fig. 6g,h), the closest being snake venom agkisacucetin (PDB code 1v4l28, 2.1 Å RMSD). TopSearch22 analysis indicates that the majority of C-type lectins use a different face for trimerization, and the only homologue to match Bd2740 is the major viral adhesion protein Mtd from Bordetella bacteriophage (PDB code 2iou29), which binds host pertactin on its upper surface. Bd2740, like agkisacucetin and Mtd, has dispensed with the Ca-binding motif (Fig. 6h; with the closest Ca-binding homologue IX-BP, PDB code 1BJ3 (ref. 30), 2.53 Å RMSD). The largest pockets and clefts are upwardly facing (distal to the fibre base) akin to Mtd. These observations reveal further potential for utilization of known adhesive folds at MAT fibre termini and show that these can diverge over time into distinct variants (Fig. 6i).
Screening of adhesin specificity for common glycans
No previous identification linked any B. bacteriovorus protein and adhesin to specific prey; we therefore assayed 6 MAT constructs (fluorescently labelled tips of Bd3182, Bd2133, Bd1334, Bd2439 and Bd2734 plus full-length Bd213321–1,031) against a commercial O-antigen array (26 glycans from 11 bacteria). Bd2439 showed strong binding to Proteus mirabilis OXK glycan, with weaker (but significant) binding to glycans from E. coli K-235 and Serratia marcescens (Fig. 6e and Supplementary Fig. 25). Our demonstration of a MAT binding pocket capable of recognizing two adjacent hydroxyls presumably explains part of this specificity, but the ubiquity of this feature in surface glycans and relative specificity of Bd2439 suggest that multiple flanking pockets will provide additional selectivity. This study has positively identified prey surface recognition proteins in predatory bacteria.
Discussion
Using independent but complementary approaches, we identified and characterized a MAT superfamily of trimeric fibre proteins with diverse adhesive tips, able to fulfil the long-sought-after role of multiplicity of molecular recognition and handling of diverse epitopes (across a wide prey range), by the bacterial predator B. bacteriovorus (Supplementary Fig. 26). The MAT family proteins fulfil the logical expectations of a surface recognition system, which are as follows: (1) surface exposed on the predator and of sufficient length to project and contact prey, (2) present in numbers that scale to prey diversity, (3) mosaic in nature in which one ‘end’ is conserved for presentation and retention on the predator and the other is diversified for different prey, (4) relatively stable and robust for the extracellular environment and (5) able to undergo dynamic re-localization associated with prey interactions during invasive predation.
We postulate that MAT proteins are dynamically expressed on the predator surface as they interact with and enter prey—in some cases at the predator’s invasive pole and in other cases at the flagellar pole (Bd1334) or midcell, even with one or two foci per family member.
MAT adhesins may fulfil several roles during prey entry, from initial attachment through dynamic relocation to porthole resealing and positional stabilization of the predatory-associated vesicle inside prey. Multiple, dynamic locations for MAT members are in keeping with the various staged processes of the Bdellovibrio life cycle that require ever-changing (but distinct) coordination between the surfaces of predator and prey (Supplementary Fig. 26).
One adhesin could recognize several prey-surface types (shared chemistry), and one prey could be recognized by several adhesins, hence the MAT family’s large redundancy and diversity to cope with a wide prey range. Thus, it is expected that the deletion of individual MAT fibre genes rarely yielded mutants significantly defective in predation. However, a slight reduction in prey entry efficiency was seen for deletion mutants of Bd2734 and Bd2740, which are structurally divergent but have similar expression profiles and are encoded at nearby genetic loci31,32. Bd2734 and Bd2740 are two of the shorter MAT fibres (843 and 627 aa, respectively (Fig. 6i)), which are relevant given that one of the short MATs we were unable to crystallize (Bd2872, 626 aa) was previously identified in a screen for predation-deficient mutants33.
As the flagellum and sheath are re-absorbed back inside the periplasm of B. bacteriovorus upon invasion6, the predatory vesicle is probably bounded by a predator-derived membrane, and reorganization at the Bdellovibrio flagellar pole is likely to be associated with the laying down of our identified vesicle (Supplementary Fig. 26). Resealing the entry port could involve PBP1b–CpoBBd0635-mediated events and would be accompanied by a septation-like membrane bifurcation event, pinching off the vesicle.
We were able to structurally characterize six separate MAT family members for B. bacteriovorus HD100, revealing that these typically large (~1,000 aa) proteins can terminate in an S74 motif characteristic of viral tailspikes14 (Bd3182), previously uncharacterized β-sandwiches (Bd2133 and Bd1334) and domains of known adhesive usage (the TNF-like domains of Bd2439 and Bd2734 and the C-type lectin fold of Bd2740). Other bacterial strains, predatory and otherwise, may use further domains still (a selection is provided in Supplementary Tables 3 and 4).
From our experimentally derived structural data on six independent MATs, plus usage of AlphaFold18 models, meandering trimerization appears to be an invariant property of the fibres, although none of the NS74 domains show strand swapping; that is, these capping regions fold independently; this independence may aid ‘tip swapping’ as predators diversify the MAT repertoire. The (predicted) N-terminal domain of the B. bacteriovorus MAT proteins is invariable in structure and would most probably sit proximal to the predator cell surface to productively position the variable C-terminal domains. The N-terminal domain has no structural homologues in the PDB but would presumably tightly bind a B. bacteriovorus surface element (whose identification awaits further study; potential interplay between adhesion and type IV pili34,35 may also be of interest). In light of our demonstration of surface localization, it is informative that the dimensions from our experimental Bd2133 electron micrographs and AlphaFold18 models are compatible with surface fibre-like features (some attached to prey) identified on tomograms6. Likewise, the features of the vesicle partly match those of a ‘bubble-like’ structure identified at the entry portal6.
We were able to confidently identify a binding partner for MAT protein Bd2439, which showed strong binding to glycans of the known prey species Proteus mirabilis36. The fact that none of the single-gene-deletion mutants were significantly defective in predation on Proteus mirabilis hints that there are multiple factors and several MAT proteins involved, but our GlcNAc-bound Bd2439 structure identifies a potential motif that may be used (in part) to select for saccharides in Proteus mirabilis and Proteus mirabilis-like glycans.
The multiplicity of the MAT family prevents exhaustive and absolute specificity determination (as suggested by our knockout phenotypes), but we use Bd2439 as a proof of principle to link to recognition of bacteria with Proteus-like O-antigen chemistries, revealing a molecular basis for B. bacteriovorus prey handling. Our observations provide a basis for future characterization and manipulation of the bacterial predator–prey relationship.
Methods
Bacterial strains and culture
B. bacteriovorus HD100, and fluorescently tagged or gene-deletion strains, were grown predatorily on stationary-phase E. coli S17-1 prey (16 h, 29 °C), in Ca–HEPES buffer (5.94 g l−1 HEPES free acid, 0.284 g l−1 calcium chloride dihydrate, pH 7.6), or on YPSC (Yeast Extract Peptone Sodium Acetate Calcium) overlay platesas plaques within a lawn of E. coli S17-1 prey, as described previously37. To provide selection for fluorescent-tag or gene-deletion constructs, 50 mg ml−1 of kanamycin sulphate was added to growth media where appropriate. E. coli S17-1 cells, for B. bacteriovorus prey or fluorescent-tagging and gene-deletion manipulations, were grown for 16 h in YT broth (5 g NaCl, 5 g Difco Yeast extract, 8 g Difco Bacto Tryptone per litre, pH adjusted to 7.5 with NaOH) at 37 °C with shaking at 200 rpm.
Plasmid and strain construction
The primers used to generate fluorescently tagged or gene-deletion strains of B. bacteriovorus are documented in Supplementary Table 5, and the plasmid constructs used are documented in Supplementary Table 6. PCR amplification was performed with Phusion polymerase (New England Biolabs) according to the manufacturer’s instructions, using primers listed in Supplementary Table 5.
Generation of markerless deletion mutants
To construct markerless gene deletions of bd1334, bd2133, bd2439, bd2734, bd2740 and bd3182, between 750 bp and 1,000 bp of DNA upstream and downstream of the gene of interest were cloned into the suicide vector pK18mobsacB by Gibson assembly38 using the NEBuilder HiFi DNA assembly cloning kit (New England BioLabs). Gene-deletion vectors were introduced into B. bacteriovorus by conjugation (using donor E. coli S17-1 strains) and subsequently cured of the donor plasmid by sucrose suicide counter-selection, resulting in the integration of the gene knockout constructs via double-crossover homologous recombination. This process is further detailed and was described previously9,39. All gene deletions were verified by Sanger sequencing. To visualize any effect of the single-gene deletions bd2734 and bd2740, during the initial stages of the predatory cycle, a C-terminal mCherry fusion to the constitutively expressed PilZ protein Bd0064 was introduced via single-crossover recombination, illuminating the B. bacteriovorus cell body, as described previously40,41,42.
Generation of Bd3182 mutation of catalytic serine S782 to alanine
The S782A mutation of Bd3182 was introduced by amplifying the gene on either side of the S74 codon and introducing the mutation in the primers (Supplementary Table 4), followed by Gibson assembly as above.
Fluorescence tagging of B. bacteriovorus gene products
Protein fluorophore tags (mCherry, mNeonGreen, mTFP or mCerulean3) were fused to the C-terminus of target genes by PCR amplification of the target gene, without its stop codon, and amplification of the fluorophore gene, followed by Gibson cloning using the NEBuilder HiFi or GeneArt assembly kits (New England Biolabs) according to the manufacturer’s instructions, into the mobilizable broad-host-range vector pK18mobsacB43. Each construct was conjugated into B. bacteriovorus HD100 as described previously37. In some cases, a particular fluorescent tag, such as mCherry for Bd2133, was not tolerated by a gene under study for reasons unknown. In such cases, a different colour combination of strains was used in experiments. Single-crossover constructs of Bd0635–mCherry and Bd0635–mTeal were obtained by standard cloning methods rather than by Gibson assembly (details in Supplementary Table 4).
For multiple fluorophore combinations, either the Bd0064 or the CpoBBd0635 tagging constructs were made with 1,000 bp of flanking DNA and ex-conjugants of these were subjected to sucrose suicide selection to generate a double-crossover event, replacing the genomic copy of the gene with the tagged version, via the same methodology used to generate single-gene-deletion strains (above). The second fluorophore-tagged gene was then introduced, where required, as above by single crossover into the B. bacteriovorus genome, as before.
Labelling of cell wall muropeptides with fluorescent d-amino acids and imaging
Pulse labelling of predator-modified prey peptidoglycan during early predation events with the ‘blue’ fluorescent d-amino acid HADA to label predatory porthole formation was carried out as described previously1. B. bacteriovorus HD100 cells were grown predatorily for 16 h at 29 °C on stationary-phase E. coli S17-1 prey, until the prey culture was lysed. The B. bacteriovorus were then filtered through a 0.45 µm filter (yielding ~2 × 108 plaque-forming units (pfu) ml−1) and concentrated 30 times by centrifugation at 12,000 × g for 5 min. The resulting pellet was resuspended in Ca–HEPES buffer (2 mM CaCl2, 25 mM HEPES, pH 7.6). E. coli S17-1 cells were grown for 16 h in Luria–Bertani medium at 37°C with shaking at 200 rpm and were back diluted to an optical density at 600 nm (OD600) of 1.0 in Ca–HEPES buffer (yielding ~1 × 109 colony-forming units (cfu) ml−1). A total of 50 μl of this B. bacteriovorus culture was mixed with 40 µl of the pre-labelled E. coli and 30 µl of Ca–HEPES buffer and incubated at 30 °C. For pulse labelling, 1.2 µl of a 50 mM stock of HADA in DMSO was added 10 min before the sampling time point for microscopy and returned to 30 °C incubation. At each time point, all of the 120 µl predator–prey sample was transferred to 175 µl ice-cold ethanol and incubated at −20 °C for at least 15 min to fix the cells. The cells were pelleted by centrifugation at 17,000 × g for 5 min, washed with 500 µl PBS and resuspended in 5 µl Slowfade (Molecular Probes), and stored at −20 °C before imaging. Samples (2 µl) were imaged using a Nikon Ti-E inverted fluorescence microscope equipped with a Plan Apo 100× 1.45 Ph3 objective, a DAPI filter cube and an Andor Neo sCMOS camera using DAPI settings for detection of HADA (emission maximum 450 nm).
B. bacteriovorus predation on E. coli in liquid culture
Assays were based on, and modified from, those detailed in a previous study11. In summary, B. bacteriovorus gene-deletion mutants were grown predatorily (as above). The pfu inputs for each gene-deletion strain were matched using the SYBR Green DNA stain, to ensure that equal titres of B. bacteriovorus for each strain were used as starting inputs. Briefly, B. bacteriovorus were incubated with SYBR Green dye for 90 min (300 rpm double orbital, in darkness, in triplicate), before fluorescence was measured using a FLUOstar Omega plate reader (BMG Labtech; excitation 485 nm, emission 520 nm, gain 800), with fluorescence values being interpolated into relative pfu per ml counts using a pfu–SYBR Green fluorescence correlation curve. E. coli S17-1 cells were grown as above and subsequently back diluted to OD600 1.0 (approximately 1 × 109 cfu ml−1) in dilute nutrient broth. Predatory cultures (containing approximately 1 × 109 cfu ml−1 E. coli prey and 1 × 108 pfu ml−1 B. bacteriovorus WT or mutant) were inoculated in triplicate into a black OptiPlate (Corning), along with media-only, B. bacteriovorus-only (no prey) and prey-only (no B. bacteriovorus) controls. The OD600 of the predatory culture was measured every 20 min, for 18 h (200 rpm, double orbital, in triplicate) to give a prey survival curve, in which a drop in OD600 is indicative of successful predation and prey lysis, because prey cells, but not predators, are large enough to produce an optical density at 600 nm.
OD600 data were exported to Excel 2016 and then analysed using CurveR, according to the method documented previously11 to analyse prey cell lysis and predation dynamics. Rmax indicates the maximum rate of prey cell lysis. S indicates the inflection point, the time at which the maximum rate of prey cell lysis (Rmax) occurs, that is, the steepest point on the curve.
To further assess the effect of deletion in early stages of the predatory cycle, synchronous predatory infections of B. bacteriovorus HD100:Bd0064mCherry:Δbd2734 and HD100: Bd0064mCherry:Δbd2740 on E. coli S17-1 PZMR100 were set up as described above, with aliquots removed at 30 min after the mixing of predator and prey. Cells were immobilized on a thin 1% Ca–HEPES buffer agarose pad and imaged as above. Images were minimally processed using the sharpen and smooth tools (ImageJ), with adjustments to brightness and contrast to ensure clarity. Full fields of view were visually scored for complete B. bacteriovorus entry as described previously44. The percentage of bdelloplasts with a fully entered B. bacteriovorus at 30 min was derived from manual inspection of the total number of bdelloplasts with B. bacteriovorus fully entered at 30 min divided by the total number of bdelloplasts visually characterized. Images were analysed from three biological repeats. Each deletion mutant strain was imaged and analysed alongside B. bacteriovorus HD100 (control) within the same experiment.
Phase-contrast and epifluorescence microscopy
The in vivo fluorescence of MAT protein–XFP tags (XFP meaning any of the different fluorescent protein tags) during the predatory life cycle was examined. Approximately synchronous predation of E. coli S17-1 PZMR100 by B. bacteriovorus strains was prepared by combining a 10-times-concentrated B. bacteriovorus predatory culture with E. coli S17-1 PZMR100 (standardized to an OD600 of 1) and Ca–HEPES at a ratio of 5:4:3, respectively, as described previously32,45. Progress through the predatory life cycle (and position of the fluorescently tagged protein under investigation) was visualized via fluorescence microscopy (10 s exposure) at the following time points (in minutes): 0, 15, 30, 45, 60, 120, 180 and 240, by withdrawing 10 μl of the culture and immobilizing on a thin 1% Ca–HEPES buffer agarose pad. Cells were visualized with a Nikon Ti-E inverted epifluorescence microscope equipped with a Plan Apo 100× Ph3 oil objective lens (Numerical Aperture 1.45) and the following filters: mCherry (excitation 550–600 nm, emission 610–665 nm), GFP, green fluorescent protein, (for mNeonGreen; excitation 460–500 nm, emission 515–530 nm), CFP, cerulean fluorescent protein, (mCerulean; excitation 420–450 nm, emission 470–490 nm) and TFP, teal fluorescent protein, (mTeal; excitation 420–450 nm, emission 515–530 nm).
Images were acquired using an Andor Neo sCMOS camera with Nikon NIS software and analysed using ImageJ (Fiji). Images were minimally processed using the sharpen and smooth tools, with adjustments to brightness and contrast. Where stated, false colour was used in channels for display purposes.
Image analysis
Images were manipulated with ImageJ (Fiji distribution) software using the sharpen and smooth tools, and by duplication of the region of interest for presentation. Images were analysed using the MicrobeJ plug-in for ImageJ46, which automates the detection of bacteria within an image. Attack-phase B. bacteriovorus cells were detected with parameters as default, with fluorescence detected by the foci method with default parameters and associated with parent bacteria with a tolerance of 0.1 µm.
For the analysis of CpoB–mCherry foci with Bd0064–mCerulean cytoplasmically labelled B. bacteriovorus, prey E. coli were detected as bacteria, B. bacteriovorus cells were detected using the medial axis method in the mCerulean channel and associated with bacteria with a tolerance of 0.1 µm, and CpoB–mCherry foci were detected using the fit shape as circle method in the mCherry channels with a maximum area of 0.25 µm2 and associated with B. bacteriovorus cells with a tolerance of 1 µm.
Manual inspection of the analysed images confirmed that the vast majority of cells and foci were correctly assigned. The shape measurements including the angularity, area, aspect ratio, circularity, curvature, length, roundness, sinuosity, solidity and width were measured for each type of cell.
Co-localization analysis
Images were analysed using ImageJ software (Fiji), using the cell counter plug-in. Adjustments to brightness and contrast for whole images were made until CpoBBd0635 (mCherry: Bd2133–mNeonGreen or mTeal (Bd2439–mCherry or Bd2740–mCherry)) and Bd2133–mNeonGreen, Bd2439–mCherry or Bd2740–mCherry foci, if present, were visible. Bdelloplasts were then manually (visually) scored for the presence of a CpoBBd0635 focus. Of the bdelloplasts that contained a CpoBBd0635 focus, the number of bdelloplasts for which a Bd2133, Bd2439 or Bd2740 focus was coincident was scored for approximately 60 bdelloplasts, originating from two biological replicates.
Visualization of external mCherry-tagged MAT protein expression
Semi-synchronous predation experiments were set up as above. Samples of 1.4 ml were pre-fixed in 0.25% paraformaldehyde and recovered by centrifugation at 17,000 × g for 2 min, then fixed in 2.5% paraformaldehyde in Dulbecco’s PBS for 10 min at 37 °C. Cells were recovered by centrifugation at 17,000 × g for 2 min, washed twice in 100 µl blocking solution (2% bovine serum albumin, BSA, in Dulbecco’s phosphate buffered saline, PBS) and then incubated in blocking solution for 45 min. Samples were further incubated in anti-mCherry antibody (Invitrogen PA5-34974) diluted 1:1,000 in blocking solution for 1 h. Cells were recovered by centrifugation at 17,000 × g for 2 min, then washed twice in blocking solution before a final incubation with secondary antibody of goat anti-rabbit IgG with Alexa Fluor plus 488 (Invitrogen A32731). Cells were then washed twice with blocking solution before imaging with a Nikon Ti-E microscope as described above. Some samples were further stained with FM46-4 by resuspension in 10 mM stain in water. Images were analysed using ImageJ software (Fiji). Adjustments to brightness and contrast for whole images were made until FM46-4-stained flagella and anti-mCherry foci were visible. Cells were then manually (visually) scored for the presence of flagella (FM46-4 red channel) and the presence and positioning of anti-mCherry foci (GFP channel).
Microscopic statistical analysis
Statistical analysis was performed in Prism 8.2.0 (GraphPad). Data were first tested for normality and then analysed using the appropriate statistical test. The number of biological repeats, n values for cell numbers and the statistical test applied are described within each figure legend.
Cloning, expression and purification
Bd213321–1,031 was synthesized and inserted into pET29a between NdeI and XhoI (Twist Bioscience), producing a construct with an N-terminal PelB leader sequence and a hexa-His tag. Bd2133662–1,031 and Bd3182668–922 were cloned into pCold1 using NdeI and XhoI restriction sites. The final constructs contained an N-terminal 24-amino-acid tag containing a transcription-enhancing element, a hexa-His tag, a factor Xa cleavage site and a Tobacco Etch Virus (TEV) cleavage site. Bd2133910–1,031 was produced by Q5 (New England Biolabs) deletion using the Bd2133662–1,031 plasmid as a template. Bd3182668–922_S782A was produced by Q5 mutagenesis using Bd3182632–922 as a template. Bd1334818–1,031 and Bd1334914–1,151 were cloned into pCold1 using XhoI and HindIII restriction sites. The final constructs contained an N-terminal 30-amino-acid tag containing a transcription-enhancing element, a hexa-His tag, a factor Xa cleavage site and a TEV cleavage site. Bd2734691–843 was cloned into pET26b using NcoI and XhoI restriction sites. The final construct contained a pelB leader sequence, Met-Ala and a hexa-His tag. Bd2439837–1,107 was cloned into pET26b using BamHI and XhoI restriction sites. The final construct contained a pelB leader sequence, Met-Asp-Ile-Phe-Ile-Asn-Ser-Asp-Pro and a hexa-His tag. Bd2740518–627 was synthesized and inserted into pET29a between NdeI and XhoI (Twist Bioscience), producing a construct with an N-terminal PelB leader sequence, a hexa-His tag and an Ala-Ser linker.
Constructs were expressed in E. coli BL21 RIPL (DE3). Cells were grown to an OD600 of 0.5–0.7 and induced with 0.5 mM IPTG (isopropyl ß-D-1-thiogalactopyranoside) at 18 °C for 16 h. Cells were harvested by centrifugation and resuspended in 50 mM HEPES, 500 mM NaCl and 20 mM imidazole, pH 7.5. The cells were lysed by sonication on ice with the exception of Bd213321–1,031, which was lysed by three passages through an EmulsiFlex C3 high-pressure homogenizer (Avestin). The lysates were loaded onto a 5 ml Ni-NTA column (GE Healthcare) and washed with 10 column volumes of lysis buffer. The protein was eluted with a gradient of 20–500 mM imidazole. TEV was added to constructs containing cleavage sites that were subsequently dialyzed into 150 mM NaCl, 20 mM HEPES and 20 mM imidazole, pH 8.0, overnight at 4 °C. The protein was then passed through a 1 ml Ni-NTA column to remove TEV and uncleaved protein. The proteins were finally passed through a Superdex 200 26/60 column (GE Healthcare), which was pre-equilibrated with 20 mM HEPES and 150 mM NaCl, pH 7.5.
To express selenomethionine-labelled Bd2133662–922, a 60 ml overnight culture of BL21 RIPL (DE3) cells in LB was centrifuged and resuspended in minimal media supplemented with kanamycin and chloramphenicol to a final volume of 1 l. At OD600 0.4, 0.1 g of lysine, 0.1 g of threonine, 0.1 g of phenylalanine, 0.05 g leucine, 0.05 g of isoleucine, 0.05 g of valine and 0.06 g of selenomethionine were added to the culture. After 15 min, IPTG was added to 1 mM. Cells were harvested by centrifugation after 16 h at 18 °C and stored at −20 °C. SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) gels were imaged using Quantity One v4.6.8.
Crystallization and data collection
For crystallization, proteins were concentrated to 3–10 mg ml−1 using a spin concentrator. Protein was mixed with precipitant in a 1:1 ratio, with drop sizes of 0.8–4 μl, and crystallized using vapour diffusion at 18 °C.
Crystals of Bd3182632–922 formed in Morpheus screen C1 (0.03 M sodium nitrate, 0.03 M sodium phosphate dibasic, 0.03 M ammonium sulphate, 0.1 M imidazole and MES (2-ethanesulfonic acid), pH 6.5, 20% v/v PEG (polyethylene glycol) 500 MME (monomethylether), 10% w/v PEG 20000) in spacegroup P21, Morpheus screen G12 (1.0 M sodium citrate tribasic dihydrate, 0.1 M HEPES, pH 7.0) with spacegroup I2 and Proplex screen E3 (0.1 M magnesium acetate tetrahydrate, 0.1 M MES, pH 6.5, 10% w/v PEG 10000) in a different P21 spacegroup. Crystals were subject to diffraction at Diamond Light Source at 100 K at wavelength 0.970–0.976 Å on beamline IO3. The phases were solved using Phenix MR47 with a low sequence homology (<10% sequence ID) ensemble of pruned T5 phage-tail fibre (PDB code 4UW8 (ref. 14)) and GP12 (PDB code 3GW6 (ref. 10)).
Crystals of Bd2734691–843 grew in JCSG+ screen A3 (0.2 M ammonium citrate dibasic, 20% w/v PEG 3350) in spacegroup P21. Crystals were subject to diffraction at Diamond Light Source at 100 K at wavelength 0.98 Å on beamline IO4. The phases were solved using Phenix MR47 with a homology model generated by ColabFold13.
Crystals of Bd1334914–1,151 formed in PACT screen B9 (0.2 M lithium chloride, 0.1 M MES, pH 6.0, 20% w/v PEG 6000) in spacegroup R3. Crystals were subject to diffraction at Diamond Light Source at 100 K at wavelength 0.98 Å on beamline IO4. Crystals of Bd1334818–1,151 grew in Morpheus screen C1 (0.03 M sodium nitrate, 0.03 M sodium phosphate dibasic, 0.03 M ammonium sulphate, 0.1 M imidazole and MES, pH 6.5, 20% v/v PEG 500 MME, 10% w/v PEG 20000) in spacegroup C2. Crystals were subject to diffraction at Diamond Light Source at 100 K at wavelength 1 Å on beamline IO4. The phases of the Bd1334818–1,151 crystals were solved using Phenix MR47 with a homology model generated by ColabFold13. The phases of the Bd1334914–1,151 crystals were solved using Phenix MR47 with a fragment of the Bd1334818–1,151 structure.
Crystals of Bd2439837–1,107 grew in multiple conditions: PACT H3 (0.2 M sodium iodide, 0.1 M Bis–Tris propane, pH 8.5, 20% w/v PEG 3350) in spacegroup P21221 and MIDASplus E9 (0.1 M lithium sulphate, 0.1 M tris, pH 8.0, 25% v/v Jeffamine ED-2003) in spacegroup P4. Crystals were cryocooled in the mother liquor with an addition of 25% ethylene glycol. Crystals were subject to diffraction at the European Synchrotron Radiation Facility at 100 K at wavelength 0.976 Å on beamlines id30a and id30b. The phases were solved using Phenix MR47 with a homology model generated by ColabFold13. Bd2439837–1,107–GlcNAc–MurNAc crystals were grown in PACT H3 as above, supplemented with 100 mM GlcNAc–MurNAc. To prevent ethylene glycol entering the binding pocket, these crystals were cryoprotected with 25% PEG 400.
Crystals of native and selenomethionine-labelled Bd2133662–1,031 grew in Morpheus screen B3 (0.03 M sodium fluoride, 0.03 M sodium bromide, 0.03 M sodium iodide, 0.1 M imidazole and MES, pH 6.5, 20% v/v glycerol, 10% w/v PEG 4000) in spacegroup P6322. Crystals were subject to diffraction at Diamond Light Source at 100 K at wavelength 0.98 Å (native) on beamline IO4-1 and 0.89 Å (selenomethionine protein) on beamline IO3. The phases of the Bd2133662–1,031 crystals were solved using experimental phasing. Selenium sites of the selenomethione-labelled protein crystals were identified via SAD by SHELX48, and Phenix47 Autosol was used to phase the data via SIRAS using the selenomethionine and native datasets. For model building, the symmetry was dropped to p63 to allow asymmetric modelling of the N-terminal beta strands. Molecular replacement of the selenomethioine-labelled protein structure was used to solve the phases of the native dataset. Crystals of Bd2133910–1,031 grew in Morpheus screen A4 (0.03 M magnesium chloride, 0.03 M calcium chloride, 0.1 M imidazole and MES, pH 6.5, 12.5% v/v MPD (2-methyl-2,4-pentanediol), 12.5% PEG 1000, 12.5% w/v PEG 3350) in spacegroup P212121. Crystals were subject to diffraction at Diamond Light Source at 100 K at wavelength 0.75 Å on beamline IO4 and solved using a fragment of the larger model.
Bd2740518–627 crystallized in MIDAS screen F1 (0.1 M HEPES, pH 6.5, 40% polypropylene glycol bisaminopropylether 2000) in space group P212121. Crystals were subject to diffraction at Diamond Light Source at 100 K at wavelength 0.979 Å on beamline IO4 and solved using molecular replacement with a ColabFold13 model.
All structures were manually completed and altered in CCP4i2 v1.1.0 and COOT 0.9.8.1 (ref. 49), and refined using Phenix 1.20.1-4487 (ref. 47). Statistics for data collection and refinement are presented in Supplementary Table 2.
Glycan arrays
Glycan arrays were performed as a service by Z Biotech. The array was blocked for 30 min using glycan array blocking buffer (Z Biotech item number 10106). Alexafluor-555-labelled samples were diluted in glycan array assay buffer (Z Biotech item number 10107) to the desired concentrations and then applied directly to the array. The array was covered with an adhesive film and shaken at 80 rpm for 1 h at room temperature. The array was then washed three times with glycan array assay buffer and two times with MilliQ water. The array was read using an Innopsys InnoScan 710 Microarray Scanner with a high-power laser at 1× photomultiplier gain. Proprietary software was used to detect each spot on the array and calculate the relative fluorescence unit (RFU) intensity for each spot. Background RFU was subtracted from each spot’s RFU value. The median of four repeat spots was determined for each glycan.
Electron microscopy
To visualize a full-length fibre, Bd213321–1,031 was subjected to negative-stain electron microscopy. Formvar and carbon grids (EMResolutions) were glow discharged with an Elmo glow discharge system (Cordouan) for 1 min at 3 mA. The protein was diluted to 1 μg ml−1 in 150 mM NaCl and 20 mM HEPES, pH 7.5, and 5 μl of protein was added followed by 5 μl of 2% uranyl acetate. Micrographs were taken using a 200 kV LaB6 Jeol 2100Plus 200 kV with a Gatan OneView IS at 50,000× magnification.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Atomic coordinates have been deposited in the PDB with accession codes 8ONC (https://doi.org/10.2210/pdb8ONC/pdb; Bd3182632–922 form 1), 8OJN (https://doi.org/10.2210/pdb8OJN/pdb; Bd3182632–922 form 2), 8ONB (https://doi.org/10.2210/pdb8ONB/pdb; Bd3182632–922 form 3), 8OND (https://doi.org/10.2210/pdb8OND/pdb; Bd2133662–1,031), 8OK3 (https://doi.org/10.2210/pdb8OK3/pdb; Bd2133910–1,031), 8OML (https://doi.org/10.2210/pdb8OML/pdb; Bd1334818–1,151), 8ON4 (https://doi.org/10.2210/pdb8ON4/pdb; Bd1334914–1151), 8OL4 (https://doi.org/10.2210/pdb8OL4/pdb; Bd2439837–1,107 with GlcNAc–MurNAc), 8ONF (https://doi.org/10.2210/pdb8ONF/pdb; Bd2439837–1,107 with ethylene glycol), 8OKW (https://doi.org/10.2210/pdb8OKW/pdb; Bd2734691–843) and 8OKS (https://doi.org/10.2210/pdb8OKS/pdb; Bd2740518–627). The mass spectrometry proteomics data have been deposited at the Oxford Research Archive (ORA) repository (https://ora.ox.ac.uk/objects/uuid:e8eff929-e8d9-4a8c-baf5-eccaf5cd7926). Molecular replacement models include the PDB 4UW8 T5 tail fibre (https://doi.org/10.2210/pdb4UW8/pdb) and the PDB 3GW6 intramolecular chaperone (https://doi.org/10.2210/pdb3GW6/pdb). Source data are provided with this paper.
References
Kuru, E. et al. Fluorescent d-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat. Microbiol. 2, 1648–1657 (2017).
Lerner, T. R. et al. Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 8, e1002524 (2012).
Lambert, C. et al. Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus. Nat. Commun. 6, 8884 (2015).
Varon, M. & Shilo, M. Interaction of Bdellovibrio bacteriovorus and host bacteria. J. Bacteriol. 95, 744–753 (1968).
Abram, D., Castro e Melo, J. & Chou, D. Penetration of Bdellovibrio bacteriovorus into host cells. J. Bacteriol. 118, 663–680 (1974).
Kaplan, M. et al. Bdellovibrio predation cycle characterized at nanometre-scale resolution with cryo-electron tomography. Nat. Microbiol. 8, 1267–1279 (2023).
Gray, A. N. et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife 4, e07118 (2015).
Lambert, C. et al. Interrupting peptidoglycan deacetylation during Bdellovibrio predator–prey interaction prevents ultimate destruction of prey wall, liberating bacterial-ghosts. Sci. Rep. 6, 26010 (2016).
Lambert, C. & Sockett, R. E. Nucleases in Bdellovibrio bacteriovorus contribute towards efficient self-biofilm formation and eradication of preformed prey biofilms. FEMS Microbiol. Lett. 340, 109–116 (2013).
Schulz, E. C. et al. Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding. Nat. Struct. Mol. Biol. 17, 210–215 (2010).
Remy, O. et al. An optimized workflow to measure bacterial predation in microplates. STAR Protoc. 3, 101104 (2022).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Garcia-Doval, C. et al. Structure of the receptor-binding carboxy-terminal domain of the bacteriophage T5 L-shaped tail fibre with and without its intra-molecular chaperone. Viruses 7, 6424–6440 (2015).
Wu, P. et al. Crystal structure of the MyRF ICA domain with its upstream β-helical stalk reveals the molecular mechanisms underlying its trimerization and self-cleavage. Int J. Biol. Sci. 17, 2931–2943 (2021).
Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022).
Steinbacher, S. et al. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl Acad. Sci. USA 93, 10584–10588 (1996).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01773-0 (2023).
Jabs, A., Weiss, M. S. & Hilgenfeld, R. Non-proline cis peptide bonds in proteins. J. Mol. Biol. 286, 291–304 (1999).
van Kempen, M. et al. Foldseek: fast and accurate protein structure search. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01773-0 (2023).
Wiederstein, M. et al. Structure-based characterization of multiprotein complexes. Structure 22, 1063–1070 (2014).
Vigolo, M. et al. A loop region of BAFF controls B cell survival and regulates recognition by different inhibitors. Nat. Commun. 9, 1199 (2018).
Shapiro, L. & Scherer, P. E. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8, 335–338 (1998).
Legrand, P. et al. The atomic structure of the phage Tuc2009 baseplate tripod suggests that host recognition involves two different carbohydrate binding modules. mBio 7, e01781-15 (2016).
Ressl, S. et al. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure 23, 688–699 (2015).
Mayer, S., Raulf, M. K. & Lepenies, B. C-type lectins: their network and roles in pathogen recognition and immunity. Histochem. Cell Biol. 147, 223–237 (2017).
Huang, K. F. et al. Crystal structure of a platelet-agglutinating factor isolated from the venom of Taiwan habu (Trimeresurus mucrosquamatus). Biochem. J. 378, 399–407 (2004).
Miller, J. L. et al. Selective ligand recognition by a diversity-generating retroelement variable protein. PLoS Biol. 6, 1195–1207 (2008).
Mizuno, H. et al. Crystal structure of coagulation factor IX-binding protein from habu snake venom at 2.6 angstrom: implication of central loop swapping based on deletion in the linker region. J. Mol. Biol. 289, 103–112 (1999).
Lambert, C. Transcriptional Analysis of Bdellovibrio bacteriovorus Responses to Different Prey and Environments. MRes thesis, Univ. Nottingham (2022).
Lambert, C. et al. The first bite—profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS ONE 5, e8599 (2010).
Duncan, M. C. et al. High-throughput analysis of gene function in the bacterial predator Bdellovibrio bacteriovorus. mBio 10, e01040-19 (2019).
Capeness, M. J. et al. Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links type IVa pilus extrusion/retraction status to prey-independent growth signalling. PLoS ONE 8, e79759 (2013).
Volle, C. B. et al. AFM force mapping elucidates pilus deployment and key lifestyle-dependent surface properties in Bdellovibrio bacteriovorus. Langmuir 39, 4233–4244 (2023).
Stolp, H. & Starr, M. P. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29, 217–248 (1963).
Lambert, C. & Sockett, R. E. Laboratory maintenance of Bdellovibrio. Curr. Protoc. Microbiol (2008). Chapter 7:Unit 7B.2. https://doi.org/10.1002/9780471729259.mc07b02s9
Gibson, D. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Harding, C. J. et al. A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus. Nat. Commun. 11, 4817 (2020).
Willis, A. R. et al. Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr. Biol. 26, 3343–3351 (2016).
Milner, D. S. et al. Ras GTPase-like protein MglA, a controller of bacterial social-motility in Myxobacteria, has evolved to control bacterial predation by Bdellovibrio. PLoS Genet. 10, e1004253 (2014).
Banks, E. J. et al. Asymmetric peptidoglycan editing generates cell curvature in Bdellovibrio predatory bacteria. Nat. Commun. 13, 1509 (2022).
Schafer, A. et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 (1994).
Banks, E. J. et al. An MltA-like lytic transglycosylase secreted by Bdellovibrio bacteriovorus cleaves the prey septum during predatory invasion. J. Bacteriol. (2023). 205(4):e0047522. doi: 10.1128/jb.00475-22
Baker, M. et al. Measuring and modelling the response of Klebsiella pneumoniae KPC prey to Bdellovibrio bacteriovorus predation, in human serum and defined buffer. Sci. Rep. 7, 8329 (2017).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016).
Zwart, P. H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Andres, D. et al. An essential serotype recognition pocket on phage P22 tailspike protein forces Salmonella enterica serovar Paratyphi A O-antigen fragments to bind as nonsolution conformers. Glycobiology 23, 486–494 (2013).
Gao, Y. X. et al. Crystal structure of agkisacucetin, a Gpib-binding snake C-type lectin that inhibits platelet adhesion and aggregation. Proteins 80, 1707–1711 (2012).
Acknowledgements
S.G.C., C.L., A.L.L., R.E.S., P.R. and J.T. were funded by the Wellcome Trust Investigator Award in Science (209437/Z/17/Z). For the purposes of open access, R.E.S. and A.L.L. have applied a CC BY public copyright licence for the author-accepted manuscript version arising from this submission. R.T. was partially funded by the same award and partly by the University of Nottingham. A.A.-B. was funded by PhD studentship from the Government of Iraq. S.G. was a self-funded University of Nottingham undergraduate student. E.J.B. and C.C. were funded by Wellcome Trust PhD studentships (215025/Z/18/Z and 220105/Z/20/Z, respectively) and University of Nottingham and Birmingham AAMR DTP Scheme 108876/Z/15/Z. E.P. was funded for this proteomic analysis by a subcontract from the Oxford Advanced Proteomics Unit, University of Oxford, UK.
Author information
Authors and Affiliations
Contributions
R.E.S. and A.L.L. jointly conceived the project and obtained funding. S.G.C. purified, crystallized and solved the structures of the proteins and investigated binding specificity with supervision from A.L.L. who identified the phage-fibre protein family in the vesicle proteome and assisted in all of the protein work. R.E.S. supervised the experimentation and helped analyse the microscopy data. C.L. purified the vesicles for proteomics and analysed the proteomic results; carried out fluorescence microscopic analysis, quantification of signals and statistics; quantified CpoB localization phenotypes; showed HADA co-localization with CpoB; carried out predation assays and analysis; and showed and quantified surface expression. J.T. performed and analysed microscopy for fluorescent strains, tested tagged fibre protein location and co-locations microscopically, and evaluated the invasion efficiency of deletion strains. P.R. designed, constructed and verified the majority of deletion and fluorescent strains, and carried out and analysed microscopy for certain fluorescent strains. S.G. made the CpoB–mCherry tag and first observed vesicles in the bdelloplast. A.A.-B. discovered the movement of CpoB–mCherry to the pole during predation by fluorescence microscopy and DAPI staining with S.G., and discovered the deposition of vesicles in the bdelloplast and their persistence in prey debris after predator exit, facilitating proteomics. R.T. constructed two deletion and three fluorescently tagged strains for the study. E.J.B. constructed the Bd2133 fluorescent tag and co-located the fluorescent protein location for Bd2133 with Bd0635, and performed informatic searches around the I888 motif in Bd2133. C.C. constructed the Bd2439 fluorescent tag and discovered that it was in the CpoBBd0635 vesicle position, and performed co-localization analyses between MAT proteins and CpoBBd0635, and quantification of the MATmCherry protein and visualization on the cell surface. E.P. conducted proteomic analyses on material purified by C.L. and provided experimental details and the rationale for protein-candidate identifications. S.G.C., C.L., R.E.S. and A.L.L. wrote the paper with contributions from the other authors. All authors approved of the final version before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Mohammed Kaplan, Abhirup Mookherjee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2, Figs. 1–26 and Tables 1–7.
Source data
Source Data Fig. 4
Unprocessed gel for Fig. 4a.
Source Data Fig. 6
Source data for Fig. 6e (glycan array).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Caulton, S.G., Lambert, C., Tyson, J. et al. Bdellovibrio bacteriovorus uses chimeric fibre proteins to recognize and invade a broad range of bacterial hosts. Nat Microbiol 9, 214–227 (2024). https://doi.org/10.1038/s41564-023-01552-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-023-01552-2
This article is cited by
-
Bdellovibrio bacteriovorus finds its prey
Nature Reviews Microbiology (2024)
-
Prey killing without invasion by Bdellovibrio bacteriovorus defective for a MIDAS-family adhesin
Nature Communications (2024)