Abstract
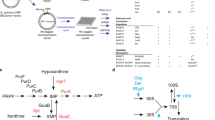
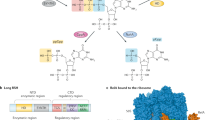
Diadenosine tetraphosphate (Ap4A) is a putative second messenger molecule that is conserved from bacteria to humans. Nevertheless, its physiological role and the underlying molecular mechanisms are poorly characterized. We investigated the molecular mechanism by which Ap4A regulates inosine-5′-monophosphate dehydrogenase (IMPDH, a key branching point enzyme for the biosynthesis of adenosine or guanosine nucleotides) in Bacillus subtilis. We solved the crystal structure of BsIMPDH bound to Ap4A at a resolution of 2.45 Å to show that Ap4A binds to the interface between two IMPDH subunits, acting as the glue that switches active IMPDH tetramers into less active octamers. Guided by these insights, we engineered mutant strains of B. subtilis that bypass Ap4A-dependent IMPDH regulation without perturbing intracellular Ap4A pools themselves. We used metabolomics, which suggests that these mutants have a dysregulated purine, and in particular GTP, metabolome and phenotypic analysis, which shows increased sensitivity of B. subtilis IMPDH mutant strains to heat compared with wild-type strains. Our study identifies a central role for IMPDH in remodelling metabolism and heat resistance, and provides evidence that Ap4A can function as an alarmone.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Structure factors and coordinates of X-ray crystallographic datasets have been deposited at the Protein Data Bank (www.rcsb.org) under the accession codes 7OJ1 and 7OJ2 for the Ap4A-bound BsIMPDH and BsIMPDH-ΔCBS, respectively. All other structural data employed in this manuscript (accession codes 3L2B, 3TSB, 4DQW, 4XTI, 4XWU, 5AHL, 5AHM, 5MCP, 6GJV, 6RPU) are publicly available in the Protein Data Bank. Source data for Figs. 1, 2, 4 and 5, and for Extended data Figs. 1, 2, 5, 6 and 8 are provided with this paper.
References
Steinchen, W. & Bange, G. The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544 (2016).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015).
Hengge, R. High-specificity local and global c-di-GMP signaling. Trends Microbiol. 29, 993–1003 (2021).
Jenal, U., Reinders, A. & Lori, C. Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017).
Stülke, J. & Krüger, L. Cyclic di-AMP signaling in bacteria. Annu. Rev. Microbiol. 74, 159–179 (2020).
Zamecnik, P. G., Stephenson, M. L., Janeway, C. M. & Randerath, K. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24, 91–97 (1966).
Ferguson, F., McLennan, A. G., Urbaniak, M. D., Jones, N. J. & Copeland, N. A. Re-evaluation of diadenosine tetraphosphate (Ap4A) from a stress metabolite to bona fide secondary messenger. Front. Mol. Biosci. 7, 332 (2020).
Despotović, D. et al. Diadenosine tetraphosphate (Ap4A) – an E. coli alarmone or a damage metabolite? FEBS J. 284, 2194–2215 (2017).
Charlier, J. & Sanchez, R. Lysyl-tRNA synthetase from Escherichia coli K12. Chromatographic heterogeneity and the lysU-gene product. Biochemical J. 248, 43–51 (1987).
Bochner, B. R., Lee, P. C., Wilson, S. W., Cutler, C. W. & Ames, B. N. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37, 225–232 (1984).
Lee, P. C., Bochner, B. R. & Ames, B. N. AppppA, heat-shock stress, and cell oxidation. Proc. Natl Acad. Sci. USA 80, 7496–7500 (1983).
Nishimura, A. et al. Diadenosine 5′,5′′′-P1,P4-tetraphosphate (Ap4A) controls the timing of cell division in Escherichia coli. Genes Cells 2, 401–413 (1997).
Farr, S. B., Arnosti, D. N., Chamberlin, M. J. & Ames, B. N. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl Acad. Sci. USA 86, 5010–5014 (1989).
Johnstone, D. B. & Farr, S. B. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10, 3897–3904 (1991).
Ji, X. et al. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc. Natl Acad. Sci. USA 116, 9578–9585 (2019).
Kimura, Y., Tanaka, C., Sasaki, K. & Sasaki, M. High concentrations of intracellular Ap4A and/or Ap5A in developing Myxococcus xanthus cells inhibit sporulation. Microbiology 163, 86–93 (2017).
Monds, R. D. et al. Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways. J. Bacteriol. 192, 3011–3023 (2010).
Lundin, A. et al. The NudA protein in the gastric pathogen Helicobacter pylori is an ubiquitous and constitutively expressed dinucleoside polyphosphate hydrolase. J. Biol. Chem. 278, 12574–12578 (2003).
Guo, W. et al. Isolation and identification of diadenosine 5′,5‴-P 1,P 4-tetraphosphate binding proteins using magnetic bio-panning. Bioorg. Med. Chem. Lett. 21, 7175–7179 (2011).
Azhar, M. A., Wright, M., Kamal, A., Nagy, J. & Miller, A. D. Biotin-c10-AppCH2ppA is an effective new chemical proteomics probe for diadenosine polyphosphate binding proteins. Bioorg. Med. Chem. Lett. 24, 2928–2933 (2014).
Fuge, E. K. & Farr, S. B. AppppA-binding protein E89 is the Escherichia coli heat shock protein ClpB. J. Bacteriol. 175, 2321–2326 (1993).
Roelofs, K. G., Wang, J., Sintim, H. O. & Lee, V. T. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl Acad. Sci. USA 108, 15528–15533 (2011).
Yang, J. et al. The nucleotide pGpp acts as a third alarmone in Bacillus, with functions distinct from those of (p) ppGpp. Nat. Commun. 11, 5388 (2020).
Hedstrom, L. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 109, 2903–2928 (2009).
Labesse, G. et al. MgATP regulates allostery and fiber formation in IMPDHs. Structure 21, 975–985 (2013).
Fernández-Justel, D., Peláez, R., Revuelta, J. L. & Buey, R. M. The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates. J. Biol. Chem. 294, 14768–14775 (2019).
Weis, D. D., Wales, T. E., Engen, J. R., Hotchko, M. & Ten Eyck, L. F. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 17, 1498–1509 (2006).
Zhang, Z. & Smith, D. L. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993).
Alexandre, T., Rayna, B. & Munier-Lehmann, H. Two classes of bacterial IMPDHs according to their quaternary structures and catalytic properties. PLoS ONE 10, e0116578 (2015).
Young, G. et al. Quantitative mass imaging of single biological macromolecules. Science 360, 423–427 (2018).
Labesse, G., Alexandre, T., Gelin, M., Haouz, A. & Munier-Lehmann, H. Crystallographic studies of two variants of Pseudomonas aeruginosa IMPDH with impaired allosteric regulation. Acta Crystallogr. D 71, 1890–1899 (2015).
Buey, R. M., Ledesma-Amaro, R., Balsera, M., de Pereda, J. M. & Revuelta, J. L. Increased riboflavin production by manipulation of inosine 5′-monophosphate dehydrogenase in Ashbya gossypii. Appl. Microbiol. Biotechnol. 99, 9577–9589 (2015).
Schäfer, H. et al. The alarmones (p)ppGpp are part of the heat shock response of Bacillus subtilis. PLoS Genet. 16, e1008275 (2020).
Lee, Y.-N., Nechushtan, H., Figov, N. & Razin, E. The function of Lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF Activity in Fc_RI-activated mast cells. Immunity 20, 145–151 (2004).
Yannay-Cohen, N. et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell 34, 603–611 (2009).
Guerra, J. et al. Lysyl-tRNA synthetase produces diadenosine tetraphosphate to curb STING-dependent inflammation. Sci. Adv. 6, eaax3333 (2020).
Luciano, D. J., Levenson-Palmer, R. & Belasco, J. G. Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol. Cell 75, 957–966.e8 (2019).
Baltzinger, M., Ebel, J. P. & Remy, P. Accumulation of dinucleoside polyphosphates in Saccharomyces cerevisiae under stress conditions. High levels are associated with cell death. Biochimie 68, 1231–1236 (1986).
Buey, R. M. et al. A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci. Rep. 7, 2648 (2017).
Buey, R. M. et al. Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat. Commun. 6, 8923 (2015).
Ereño-Orbea, J., Oyenarte, I. & Martínez-Cruz, L. A. CBS domains: ligand binding sites and conformational variability. Arch. Biochem. Biophys. 540, 70–81 (2013).
Anashkin, V. A., Baykov, A. A. & Lahti, R. Enzymes regulated via cystathionine β-synthase domains. Biochemistry 82, 1079–1087 (2017).
Kriel, A. et al. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell 48, 231–241 (2012).
Krásný, L. & Gourse, R. L. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23, 4473–4483 (2004).
Plateau, P., Fromant, M., Kepes, F. & Blanquet, S. Intracellular 5′,5′-dinucleoside polyphosphate levels remain constant during the Escherichia coli cell cycle. J. Bacteriol. 169, 419–422 (1987).
Coste, H., Brevet, A., Plateau, P. & Blanquet, S. Non-adenylylated bis(5′-nucleosidyl) tetraphosphates occur in Saccharomyces cerevisiae and in Escherichia coli and accumulate upon temperature shift or exposure to cadmium. J. Biol. Chem. 262, 12096–12103 (1987).
Fung, D. K., Yang, J., Stevenson, D. M., Amador-Noguez, D. & Wang, J. D. Small alarmone synthetase SasA expression leads to concomitant accumulation of pGpp, ppApp, and AppppA in Bacillus subtilis. Front. Microbiol. 11, 2083 (2020).
Konkol, M. A., Blair, K. M. & Kearns, D. B. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 195, 4085–4093 (2013).
Osorio-Valeriano, M. et al. ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell 179, 1512–1524.e15 (2019).
Wales, T. E., Fadgen, K. E., Gerhardt, G. C. & Engen, J. R. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 80, 6815–6820 (2008).
Geromanos, S. J. et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695 (2009).
Li, G.-Z. et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 (2009).
Mueller, U. et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: current status and perspectives. Eur. Phys. J. 130, 141 (2015).
Powell, H. R., Battye, T. G. G., Kontogiannis, L., Johnson, O. & Leslie, A. G. W. Integrating macromolecular X-ray diffraction data with the graphical user interface iMosflm. Nat. Protoc. 12, 1310–1325 (2017).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
The PyMOL Molecular Graphics System Version 1.8 (Schrödinger, 2015).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Burby, P. E. & Simmons, L. A. CRISPR/Cas9 editing of the Bacillus subtilis genome. Bio Protoc. 7, e2272 (2017).
Tuominen, H. et al. Crystal structures of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP, and activator, diadenosine tetraphosphate. J. Mol. Biol. 398, 400–413 (2010).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Makowska-Grzyska, M. et al. Bacillus anthracis inosine 5′-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes. Biochemistry 51, 6148–6163 (2012).
Alexandre, T. et al. First-in-class allosteric inhibitors of bacterial IMPDHs. Eur. J. Med. Chem. 167, 124–132 (2019).
Acknowledgements
This work was supported by the priority programme of the Deutsche Forschungsgemeinschaft (DFG) SPP1879 – ‘Nucleotide second messenger signalling in bacteria’ (to G.B.), the USA National Institute of Health R35 GM127088 and the Howard Hughes Medical Institute Faculty Scholars Award (to J.D.W.), and the National Science Foundation (NSF) grant award no. 1715710 (to D.A.-N.). We thank the European Synchrotron Radiation Facility (Grenoble, France) and the Electron Storage Ring BESSY II (Berlin, Germany) for the excellent beamline support. We acknowledge support from the ‘DFG-core facility for interactions, dynamics and macromolecular assembly structure’ at the Philipps-University Marburg (to G.B.). G.H. acknowledges support from the Free-floater programme of the Max Planck Society.
Author information
Authors and Affiliations
Contributions
J.D.W. and G.B. conceptualized the project. G.H. and D.A.-N. developed the methodology. P.I.G., M.K.M.Y., W.S., C.-N.M., J.Y. and A.P. conducted the experimental investigations. J.D.W. and G.B. acquired funding. J.D.W. and G.B. supervised the project. P.I.G., M.Y., W.S., J.D.W. and G.B. wrote the original draft. All authors read and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Natalia Tschowri and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
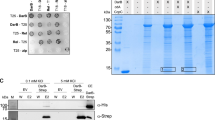
Extended Data Fig. 1 Binding of nucleotides and dinucleotides to BsIMPDH determined by isothermal titration calorimetry (ITC).
a-j. A 25 µM concentrated solution of purified BsIMPDH was titrated with a. AMP, b. ADP, c. ATP, d. Ap3A, e. Ap4A, f. Ap5A, g. Ap6A, h. Ap3G, i. Ap4G, or j. Ap5G. The differential power (DP, upper plot) for each injection of ligand was recorded and used to determine the dissociation constant Kd from a fitting of the calculated binding enthalpies (ΔH, lower plot).
Extended Data Fig. 2 Enzyme kinetic parameters obtained for BsIMPDH.
a-c. The maximal velocity (Vmax) and Michaelis-Menten constant (Km) of BsIMPDH activity were determined in the presence of various ligands. For Ap4A, the inhibitory constant (Ki) was fitted from the change in Vmax in the presence of different Ap4A concentrations. The mean ± SD of enzymological parameters was obtained with Graph Pad Prism from n = 2 replicates. For assay results displayed in a (related to Fig. 1e–h) and c (related to Extended Data Fig. 6a), the concentration of NAD+ was kept constant at 5 mM and the concentration of IMP variable between 25 and 1,000 µM. For the assay results displayed in b (related to Fig. 1e), the concentration of IMP was kept constant at 3 mM and the concentration of NAD+ variable between 25 and 5,000 µM. n.d., not determined.
Extended Data Fig. 3 Coordination of Ap4A by the BsIMPDH CBS domains.
a. The CBS domains of the B. subtilis IMPDH (red, this study) were aligned with the CBS domains of P. aeruginosa IMPDH (left; pink, PDB-ID: 4DQW25), A. gossypii IMPDH (middle; light blue, PDB-ID: 6RPU26), and C. perfringens pyrophosphatase (right; salmon, PDB-ID: 3L2B63, revealing on overall similar topology. b. The unbiased Fobs-Fcalc difference electron density map (top) is coloured in green and red for positive and negative electron density, respectively, and contoured at 3.0 σ. The Ap4A ligand was not present during the refinement. The 2Fobs-Fcalc electron density map after final refinement (bottom), including the Ap4A ligand, is coloured in blue and contoured at 1.5 σ. c. Coordination of Ap4A in chains A and B of the crystal structure of Ap4A-bound BsIMPDH. Atoms are displayed as spheres and coloured as follows: carbon, black; oxygen, red; nitrogen, blue; phosphor, purple. Purple and orange solid lines illustrate ligand or non-ligand covalent bonds, and green dashed lines represent hydrogen bonds. Red semicircles denote hydrophobic interactions. The image was generated with LigPlot+64. d. Coordination of ligands by the CBS domains of B. subtilis IMPDH with Ap4A (left), P. aeruginosa IMPDH with 2 ATP (middle, PDB-ID: 4DQW25), and A. gossypii IMPDH with Ap5G (right, PDB-ID: 6RPU26). Ligands and amino acid residues lining the ligand-binding sites are shown as sticks and coloured by element (carbon, green/yellow/cyan; oxygen, red; nitrogen, blue; phosphor, orange). Green and purple spheres represent magnesium and manganese ions, respectively. Dashed lines denote hydrogen bonding interactions between the main chain carbonyl/amide groups and the adenine and guanine nucleobases. A. gossypii IMPDH does not require a divalent metal ion to coordinate the GDP ligand, whereas metal ion coordination in the prokaryotic B. subtilis and P. aeruginosa IMPDHs is achieved by E184 and E180 residues, respectively40.
Extended Data Fig. 4 Conformational flexibility of CBS domains in crystal structures of selected IMPDH proteins.
a-f. The crystal structures of IMPDH proteins from a. B. anthracis in apo-state (PDB-ID: 3TSB65), b. B. subtilis bound to Ap4A (this study), c. P. aeruginosa in apo-state (PDB-ID: 6GJV66), d. P. aeruginosa bound to ATP (PDB-ID: 4DQW25), e. A. gossypii bound to Ap5G and GDP (PDB-ID: 6RPU26), and f. A. gossypii bound to ATP (PDB-ID: 5MCP39) are shown as ribbon and colored to their B factors from 20 (blue) to 150 Å2 (red). For octameric biological assemblies, the bottom tetrameric ring is shown in grey surface. CBS domains are indicated by black dashed rectangles, and teal spheres denote the nucleotide ligands coordinates within the CBS domains.
Extended Data Fig. 5 Influence of Ap4A and ATP on conformational flexibility of BsIMPDH CBS domains by hydrogen/deuterium exchange mass spectrometry.
a. Difference in hydrogen/deuterium exchange (HDX) of representative peptides in the presence of Ap4A (top) or ATP (bottom), expressed as the difference in HDX of ligand-bound BsIMPDH versus its apo-state. b. Location of selected peptides exhibiting bimodality in HDX (EX1 kinetics) displayed on the adjacent CBS domains of two monomers of Ap4A-bound BsIMPDH. c-d. Mass spectra of two selected BsIMPDH peptides exhibiting EX1 kinetics for hydrogen/deuterium exchange, that is, c. the peptides containing residues 113–136, and d. residues 172–200 of BsIMPDH. The occurrence of a fast-exchanging population in the apo-state (red) is indicative for unfolding of secondary structures, which is restricted in samples of Ap4A-bound (blue) and ATP-bound (green) BsIMPDH.
Extended Data Fig. 6 Enzymatic activity and oligomeric state of BsIMPDH-WT and CBS domain variants.
a. Representative distributions of tetrameric and octameric BsIMPDH species for BsIMPDH-WT in the absence of IMP and NAD+ substrates and in dependence of Ap4A or ATP concentration. b-e. Representative distributions of tetrameric and octameric BsIMPDH species of b. BsIMPDH-WT, c. BsIMPDH-K202A, d. BsIMPDH-R141A/R144A, and e. BsIMPDH-ΔCBS, all in the presence of IMP and NAD+ substrates and in dependence of Ap4A or ATP concentration. f. Representative distributions of tetrameric and octameric BsIMPDH species for BsIMPDH-WT in the presence of IMP and NAD+ substrates and in dependence of the indicated adenosine nucleotides or ApnA and ApnG dinucleotides added. In a-f, the numbers in the diagrams reflect the molecular weight of the observed oligomeric species (mean and SD), the number of observed molecules for either species and their percentage of all observed molecules in the sample. g. Effect of 10 µM Ap4A or 2 mM ATP on the enzyme-kinetic behavior of BsIMPDH-WT and CBS domain variants K202A, R141A/R144A and ΔCBS for conversion of the IMP substrate into XMP. Individual data points of n = 2 replicates are shown, and parameters of the fits are given in Extended Data Fig. 2c. h. Correlation between observed fraction of IMPDH monomers in oligomeric state and the apparent Vmax of IMPDH activity. Linear regression was performed either with values for BsIMPDH WT only (red trace) or all depicted values including WT and variants (blue trace). Red and blue numbers denote values for the fraction and Vmax at the intersects with the x-axis and y-axis, respectively.
Extended Data Fig. 7 Different conformations of catalytic flap and C-termini regions in crystal structures of IMPDH enzymes.
a-f. Cartoon representations of the crystal structures of a. Ap4A-bound full-length B. subtilis (Bs) IMPDH, b. B. subtilis (Bs) IMPDH-ΔCBS, c. P. aeruginosa (Pa) IMPDH-ΔCBS (PDB-ID: 5AHL31), d. P. aeruginosa (Pa) IMPDH-ΔCBS with substrate IMP in the active site (PDB-ID: 5AHM31), e. A. gossypii (Ag) IMPDH-ΔCBS (PDB-ID: 4XWU32), and f. A. gossypii (Ag) IMPDH-ΔCBS with substrate IMP in the active site (PDB-ID: 4XTI32). The catalytic cysteine (green), catalytic flap (red) and C-termini (orange) where colored and denoted where possible.
Extended Data Fig. 8 Quantification of rising intracellular Ap4A concentrations in B. subtilis after non-lethal heat shock.
In vivo quantification of Ap4A in wildtype (WT) B. subtilis 3610 and IMPDH mutant strains. Data represent mean ± SD of n = 3 biological replicates. Unpaired two-tailed t-tests were used to compare Ap4A levels for WT and IMPDH mutant strains of heat-shocked conditions (15 min, 30 min) versus the untreated control (0 min). Asterisks indicate p-values: * p ≤ 0.05; ns, not significant. Exact p-values are, WT: 0.1382 (15 vs. 0 min), 0.0943 (30 vs. 15 min), 0.0235 (30 vs. 0 min); K202A: 0.0191 (15 vs. 0 min), 0.5491 (30 vs. 15 min), 0.0223 (30 vs. 0 min); R141A/R144A: 0.4789 (15 vs. 0 min), 0.8778 (30 vs. 15 min), 0.3756 (30 vs. 0 min); ΔCBS: 0.0753 (15 vs. 0 min), 0.1146 (30 vs. 15 min), 0.0390 (30 vs. 0 min).
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Source data
Source Data Fig. 1
Statistical source data – numerical raw data of experiment(s).
Source Data Fig. 2
Statistical source data – numerical raw data of experiment(s).
Source Data Fig. 4
Statistical source data – numerical raw data of experiment(s).
Source Data Fig. 5
Statistical source data – numerical raw data of experiment(s).
Source Data Extended Data Fig. 1
Statistical source data – numerical raw data of experiment(s).
Source Data Extended Data Fig. 2
Statistical source data – numerical raw data of experiment(s).
Source Data Extended Data Fig. 5
Statistical source data – numerical raw data of experiment(s).
Source Data Extended Data Fig. 6
Statistical source data – numerical raw data of experiment(s).
Source Data Extended Data Fig. 8
Statistical source data – numerical raw data of experiment(s).
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Giammarinaro, P.I., Young, M.K.M., Steinchen, W. et al. Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis. Nat Microbiol 7, 1442–1452 (2022). https://doi.org/10.1038/s41564-022-01193-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01193-x