Abstract
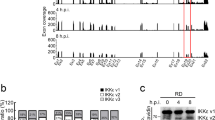
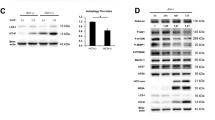
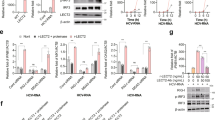
Current models of cell-intrinsic immunity to RNA viruses centre on virus-triggered inducible antiviral responses initiated by RIG-I-like receptors or Toll-like receptors that sense pathogen-associated molecular patterns, and signal downstream through interferon regulatory factors (IRFs), transcription factors that induce synthesis of type I and type III interferons1. RNA viruses have evolved sophisticated strategies to disrupt these signalling pathways and evade elimination by cells, attesting to their importance2. Less attention has been paid to how IRFs maintain basal levels of protection against viruses. Here, we depleted antiviral factors linked to RIG-I-like receptor and Toll-like receptor signalling to map critical host pathways restricting positive-strand RNA virus replication in immortalized hepatocytes and identified an unexpected role for IRF1. We show that constitutively expressed IRF1 acts independently of mitochondrial antiviral signalling (MAVS) protein, IRF3 and signal transducer and activator of transcription 1 (STAT1)-dependent signalling to provide intrinsic antiviral protection in actinomycin D-treated cells. IRF1 localizes to the nucleus, where it maintains the basal transcription of a suite of antiviral genes that protect against multiple pathogenic RNA viruses, including hepatitis A and C viruses, dengue virus and Zika virus. Our findings reveal an unappreciated layer of hepatocyte-intrinsic immunity to these positive-strand RNA viruses and identify previously unrecognized antiviral effector genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and in its Supplementary Information. The RNA-seq data have been deposited with the Gene Expression Omnibus (GSE114916).
References
Yoneyama, M., Onomoto, K., Jogi, M., Akaboshi, T. & Fujita, T. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 32, 48–53 (2015).
Chan, Y. K. & Gack, M. U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 14, 360–373 (2016).
Li, K., Chen, Z., Kato, N., Gale, M. Jr. & Lemon, S. M. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-β production in hepatocytes. J. Biol. Chem. 280, 16739–16747 (2005).
Woodson, S. E. & Holbrook, M. R. Infection of hepatocytes with 17-D vaccine-strain yellow fever virus induces a strong pro-inflammatory host response. J. Gen. Virol. 92, 2262–2271 (2011).
Feng, H. et al. NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR. Nat. Immunol. 18, 1299–1309 (2017).
Lemon, S. M., Ott, J. J., Van Damme, P. & Shouval, D. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 68, 167–184 (2018).
Hirai-Yuki, A. et al. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353, 1541–1545 (2016).
Taki, S. et al. Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity 6, 673–679 (1997).
White, L. C. et al. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+T cells in IRF-1-/-mice. Immunity 5, 365–376 (1996).
Fujita, T., Kimura, Y., Miyamoto, M., Barsoumian, E. L. & Taniguchi, T. Induction of endogenous IFN-α and IFN-β genes by a regulatory transcription factor, IRF-1. Nature 337, 270–272 (1989).
Odendall, C. et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15, 717–726 (2014).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Dixit, E. et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 (2010).
Sumpter, R.Jr et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79, 2689–2699 (2005).
Leblanc, J. F., Cohen, L., Rodrigues, M. & Hiscott, J. Synergism between distinct enhanson domains in viral induction of the human beta interferon gene. Mol. Cell. Biol. 10, 3987–3993 (1990).
Miyamoto, M. et al. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell 54, 903–913 (1988).
Tanaka, N., Kawakami, T. & Taniguchi, T. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell. Biol. 13, 4531–4538 (1993).
Oikawa, T. et al. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nat. Commun. 6, 8070 (2015).
Uyama, T., Jin, X. H., Tsuboi, K., Tonai, T. & Ueda, N. Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim. Biophys. Acta 1791, 1114–1124 (2009).
Staring, J. et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 541, 412–416 (2017).
Hsu, T. H. et al. Involvement of RARRES3 in the regulation of Wnt proteins acylation and signaling activities in human breast cancer cells. Cell Death Differ. 22, 801–814 (2015).
Ou, C. C. et al. Downregulation of HER2 by RIG1 involves the PI3K/Akt pathway in ovarian cancer cells. Carcinogenesis 29, 299–306 (2008).
Chiang, G. G. & Abraham, R. T. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 280, 25485–25490 (2005).
Figueiredo, V. C., Markworth, J. F. & Cameron-Smith, D. Considerations on mTOR regulation at serine 2448: implications for muscle metabolism studies. Cell. Mol. Life Sci. 74, 2537–2545 (2017).
Wang, J. et al. Negative regulation of Nmi on virus-triggered type I IFN production by targeting IRF7. J. Immunol. 191, 3393–3399 (2013).
McCarthy, M. K. & Weinberg, J. B. The immunoproteasome and viral infection: a complex regulator of inflammation. Front. Microbiol. 6, 21 (2015).
Verweij, M. C. et al. Viral inhibition of the transporter associated with antigen processing (TAP): a striking example of functional convergent evolution. PLoS Pathog. 11, e1004743 (2015).
Schoggins, J. W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014).
Langlais, D., Barreiro, L. B. & Gros, P. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med. 213, 585–603 (2016).
Nair, S., Poddar, S., Shimak, R. M. & Diamond, M. S. Interferon regulatory factor-1 (IRF-1) protects against chikungunya virus induced immunopathology by restricting infection in muscle cells. J. Virol. 91, e01419-17 (2017).
Dansako, H. et al. Class A scavenger receptor 1 (MSR1) restricts hepatitis C virus replication by mediating toll-like receptor 3 recognition of viral RNAs produced in neighboring cells. PLoS Pathog. 9, e1003345 (2013).
Yamane, D. et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat. Med. 20, 927–935 (2014).
Yamane, D. et al. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3′ variants. Nucleic Acids Res. 45, 4743–4755 (2017).
Feng, Z. et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371 (2013).
Hishiki, T. et al. Interferon-mediated ISG15 conjugation restricts dengue virus 2 replication. Biochem. Bioph. Res. Co. 448, 95–100 (2014).
Beard, M. R., Cohen, L., Lemon, S. M. & Martin, A. Characterization of recombinant hepatitis A virus genomes containing exogenous sequences at the 2A/2B junction. J. Virol. 75, 1414–1426 (2001).
Binn, L. N. et al. Primary isolation and serial passage of hepatitis A virus strains in primate cell cultures. J. Clin. Microbiol. 20, 28–33 (1984).
Matsuda, M. et al. High-throughput neutralization assay for multiple flaviviruses based on single-round infectious particles using dengue virus type 1 reporter replicon. Sci. Rep. 8, 16624 (2018).
Yi, M. & Lemon, S. M. Replication of subgenomic hepatitis A virus RNAs expressing firefly luciferase is enhanced by mutations associated with adaptation of virus to growth in cultured cells. J. Virol. 76, 1171–1180 (2002).
Baba, T. et al. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J. Biol. Chem. 289, 11497–11511 (2014).
Imae, R. et al. LYCAT, a homologue of C. elegans acl-8, acl-9, and acl-10, determines the fatty acid composition of phosphatidylinositol in mice. J. Lipid Res. 53, 335–347 (2012).
Kielkowska, A. et al. A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv. Biol. Regul. 54, 131–141 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
The authors thank C.M. Rice, S. Inoue and N. Kato for the reagents, M. Soloway for the bioinformatics support, M. Chua for technical assistance and H. Dansako, B.B. Queliconi and W.J. Zuercher for helpful discussions. This work was supported in part by the Japan Society for the Promotion of Science (JSPS KAKENHI) (grant nos. JP16H07462 and JP17H05070 to D.Y., grant no. JP18K05987 to A.H-Y., grant no. JP17K08870 and JP15K19109 to T.H.), Japan Agency for Medical Research and Development (grant no. JP18jk0210014 to A.H.-Y., grant no. JP18fk0108035 to T.H. and grant no. JP16fk0210108 to M.K.) and National Institutes of Health grant no. R01-AI103083 and U19-AI109965 to S.M.L. and grant no. R01-AI131685 to S.M.L. and J.K.W.
Author information
Authors and Affiliations
Contributions
D.Y. and S.M.L. conceived the study and wrote the manuscript. D.Y., H.F., E.E.R.-S., A.H.-Y., K.L.M., I.M., L.H., W.L. and O.G.-L. performed the experiments. S.R.S. and P.S. performed the bioinformatics analysis. H.N. and T.O.-N. performed the lipidomics analysis. A.D., R.S., M.M., T.H., E.W., T.O., K.M., L.M.R., M.K. and J.K.W. provided the research materials and supervised the experiments. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–10 and Supplementary Tables 1–9.
Rights and permissions
About this article
Cite this article
Yamane, D., Feng, H., Rivera-Serrano, E.E. et al. Basal expression of interferon regulatory factor 1 drives intrinsic hepatocyte resistance to multiple RNA viruses. Nat Microbiol 4, 1096–1104 (2019). https://doi.org/10.1038/s41564-019-0425-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0425-6
This article is cited by
-
Cooperative effects of RIG-I-like receptor signaling and IRF1 on DNA damage-induced cell death
Cell Death & Disease (2022)
-
N6-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA
Nature Communications (2021)
-
Constitutive immune mechanisms: mediators of host defence and immune regulation
Nature Reviews Immunology (2021)
-
Hepatocytes trap and silence coxsackieviruses, protecting against systemic disease in mice
Communications Biology (2020)