Abstract
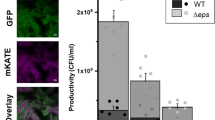
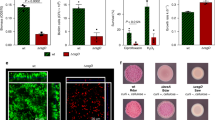
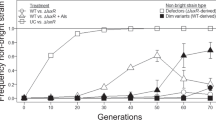
Closely related microorganisms often cooperate, but the prevalence and stability of cooperation between different genotypes remain debatable. Here, we track the evolution of pellicle biofilms formed through genetic division of labour and ask whether partially deficient partners can evolve autonomy. Pellicles of Bacillus subtilis rely on an extracellular matrix composed of exopolysaccharide (EPS) and the fibre protein TasA. In monocultures, ∆eps and ∆tasA mutants fail to form pellicles, but, facilitated by cooperation, they succeed in co-culture. Interestingly, cooperation collapses on an evolutionary timescale and ∆tasA gradually outcompetes its partner ∆eps. Pellicle formation can evolve independently from division of labour in ∆eps and ∆tasA monocultures, by selection acting on the residual matrix component, TasA or EPS, respectively. Using a set of interdisciplinary tools, we unravel that the TasA producer (∆eps) evolves via an unconventional but reproducible substitution in TasA that modulates the biochemical properties of the protein. Conversely, the EPS producer (ΔtasA) undergoes genetically variable adaptations, all leading to enhanced EPS secretion and biofilms with different biomechanical properties. Finally, we revisit the collapse of division of labour between Δeps and ΔtasA in light of a strong frequency versus exploitability trade-off that manifested in the solitarily evolving partners. We propose that such trade-off differences may represent an additional barrier to evolution of division of labour between genetically distinct microorganisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data sets generated and analysed during this study are available from the corresponding author on request.
Change history
11 January 2019
In the version of this Article originally published, author Carolina Falcón Garcia’s name was coded wrongly, resulting in it being incorrect when exported to citation databases. This has now been corrected, though no visible changes will be apparent.
References
Hamilton, W. D. The genetical evolution of social behaviour. I. J. Theor. Biol. 7, 1–16 (1964).
Queller, D. C., Ponte, E., Bozzaro, S. & Strassmann, J. E. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science 299, 105–106 (2003).
Gibbs, K. A. & Greenberg, E. P. Territoriality in Proteus: advertisement and aggression. Chem. Rev. 111, 188–194 (2011).
Kiers, T. E., Rousseau, R. A., West, S. A. & Denison, R. F. Host sanctions and the legume–rhizobium mutualism. Nature 425, 78–81 (2003).
West, S. A., Griffin, A. S. & Gardner, A. Evolutionary explanations for cooperation. Curr. Biol. 17, R661–R672 (2007).
Zhang, Z., Claessen, D. & Rozen, D. E. Understanding microbial divisions of labor. Front. Microbiol. 7, 2070 (2016).
van Gestel, J., Vlamakis, H. & Kolter, R. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol. 13, e1002141 (2015).
Dragoš, A. et al. Division of labor during biofilm matrix production. Curr. Biol. 28, 1903–1913 (2018).
Pande, S. & Kost, C. Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol. 25, 349–361 (2017).
Kim, W., Levy, S. B. & Foster, K. R. Rapid radiation in bacteria leads to a division of labour. Nat. Commun. 7, 10508 (2016).
Germerodt, S. et al. Pervasive selection for cooperative cross-feeding in bacterial communities. PLoS Comput. Biol. 12, e1004986 (2016).
Lowery, N. V., McNally, L., Ratcliff, W. C. & Brown, S. P. Division of labor, bet hedging, and the evolution of mixed biofilm investment strategies. mBio 8, e00672-17 (2017).
Wahl, L. M. The division of labor: genotypic versus phenotypic specialization. Am. Nat. 160, 135–145 (2002).
D’Souza, G. & Kost, C. Experimental evolution of metabolic dependency in bacteria. PLoS Genet. 12, e1006364 (2016).
Morris, J. J., Lenski, R. E. & Zinser, E. R. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3, e00036-12 (2012).
Oliveira, N. M., Niehus, R. & Foster, K. R. Evolutionary limits to cooperation in microbial communities. Proc. Natl Acad. Sci. USA 111, 17941–17946 (2014).
Cooper, G. A. & West, S. A. Division of labour and the evolution of extreme specialization. Nat. Ecol. Evol. 2, 1161–1167 (2018).
Waite, A. J. & Shou, W. Adaptation to a new environment allows cooperators to purge cheaters stochastically. Proc. Natl Acad. Sci. USA 109, 19079–19086 (2012).
Martin, M. et al. De novo evolved interference competition promotes the spread of biofilm defectors. Nat. Commun. 8, 15127 (2017).
O’Brien, S., Luján, A. M., Paterson, S., Cant, M. A. & Buckling, A. Adaptation to public goods cheats in Pseudomonas aeruginosa. Proc. Biol. Sci. 284, 20171089 (2017).
Kümmerli, R. et al. Co-evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa. J. Evol. Biol. 28, 2264–2274 (2015).
Fiegna, F., Yu, Y.-T. N., Kadam, S. V. & Velicer, G. J. Evolution of an obligate social cheater to a superior cooperator. Nature 441, 310–314 (2006).
Hammerschmidt, K., Rose, C. J., Kerr, B. & Rainey, P. B. Life cycles, fitness decoupling and the evolution of multicellularity. Nature 515, 75–79 (2014).
Dragoš, A. et al. Evolution of exploitative interactions during diversification in Bacillus subtilis biofilms. FEMS Microbiol. Ecol. 94, fix155 (2018).
Branda, S. S., Gonzalez-Pastor, J. E., Ben-Yehuda, S., Losick, R. & Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl Acad. Sci. USA 98, 11621–11626 (2001).
Branda, S. S., Chu, F., Kearns, D. B., Losick, R. & Kolter, R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59, 1229–1238 (2006).
Romero, D., Vlamakis, H., Losick, R. & Kolter, R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol. Microbiol. 80, 1155–1168 (2011).
Romero, D., Aguilar, C., Losick, R. & Kolter, R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl Acad. Sci. USA 107, 2230–2234 (2010).
Dragoš, A., Kovács, Á. T. & Claessen, D. The role of functional amyloids in multicellular growth and development of Gram-positive bacteria. Biomolecules 7, E60 (2017)..
Diehl, A. et al. Structural changes of TasA in biofilm formation of Bacillus subtilis. Proc. Natl Acad. Sci. USA 115, 3237–3242 (2018).
Arnaouteli, S. et al. Bifunctionality of a biofilm matrix protein controlled by redox state. Proc. Natl Acad. Sci. USA 114, E6184–E6191 (2017).
Kim, W., Racimo, F., Schluter, J., Levy, S. B. & Foster, K. R. Importance of positioning for microbial evolution. Proc. Natl Acad. Sci. USA 111, E1639–E1647 (2014).
West, S. A., Griffin, A. S. & Gardner, A. Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J. Evol. Biol. 20, 415–432 (2007).
Foster, K. R. & Bell, T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850 (2012).
Abrudan, M. I. et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl Acad. Sci. USA 112, 11054–11059 (2015).
Rainey, P. B. & Rainey, K. Evolution of cooperation and conflict in experimental bacterial populations. Nature 425, 72–74 (2003).
Scanlan, P. D. & Buckling, A. Co-evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J. 6, 1148–1158 (2012).
Miskinyte, M. et al. The genetic basis of Escherichia coli pathoadaptation to macrophages. PLoS Pathog. 9, e1003802 (2013).
Rainey, P. B. & Travisano, M. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72 (1998).
Konkol, M. A., Blair, K. M. & Kearns, D. B. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 195, 4085–4093 (2013).
Borkar, S. G. Laboratory Techniques in Plant Bacteriology (CRC Press, Boca Raton, 2017).
Werb, M. et al. Surface topology affects wetting behavior of Bacillus subtilis biofilms. NPJ Biofilms Microbiomes 3, 11 (2017).
Acknowledgements
We thank S. West for his comments on our manuscript. This work was funded by the Deutsche Forschungsgemeinschaft (DFG) to Á.T.K. (KO4741/2.1) within the Priority Program SPP1617. A.D. and C.F.G. were supported by fellowships from the Alexander von Humboldt Foundation and Consejo Nacional de Ciencia y Tecnología (CONACyT), respectively. The research leading to these results has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 713683 (COFUNDfellowsDTU). This work was also supported by the DFG through project B11 in the framework of SFB863 granted to O.L., and a start-up grant from the Technical University of Denmark to Á.T.K.
Author information
Authors and Affiliations
Contributions
A.D. and Á.T.K. conceived the project. A.D., M.M., C.F.G., L.K., P.P. and T.H. performed the experiments. B.B. performed the next-generation sequencing data analysis. G.M., G.B., D.L. and O.L. contributed with the methods, analysed the data and supervised the experiments. A.D. and Á.T.K. wrote the manuscript, with all authors contributing to the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, Supplementary References, Supplementary Figures 1–11, Supplementary Results.
Supplementary Dataset 1
List of mutations detected in evolved single isolates and populations.
Supplementary Video 1
Effects of cysteine-containing TasA on wetting behaviour of B. subtilis pellicles.
Rights and permissions
About this article
Cite this article
Dragoš, A., Martin, M., Falcón García , C. et al. Collapse of genetic division of labour and evolution of autonomy in pellicle biofilms. Nat Microbiol 3, 1451–1460 (2018). https://doi.org/10.1038/s41564-018-0263-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0263-y
This article is cited by
-
Adaptation and phenotypic diversification of Bacillus thuringiensis biofilm are accompanied by fuzzy spreader morphotypes
npj Biofilms and Microbiomes (2022)
-
Complex extracellular biology drives surface competition during colony expansion in Bacillus subtilis
The ISME Journal (2022)
-
The evolution of mechanisms to produce phenotypic heterogeneity in microorganisms
Nature Communications (2022)
-
Resistance evolution can disrupt antibiotic exposure protection through competitive exclusion of the protective species
The ISME Journal (2022)
-
Donor-strand exchange drives assembly of the TasA scaffold in Bacillus subtilis biofilms
Nature Communications (2022)