Abstract
As a conserved pathway that lies at the intersection between host defence and cellular homeostasis, autophagy serves as a rheostat for immune reactions. In particular, autophagy suppresses excess type I interferon (IFN-I) production in response to viral nucleic acids. It is unknown how this function of autophagy relates to the intestinal barrier where host–microbe interactions are pervasive and perpetual. Here, we demonstrate that mice deficient in autophagy proteins are protected from the intestinal bacterial pathogen Citrobacter rodentium in a manner dependent on IFN-I signalling and nucleic acid sensing pathways. Enhanced IFN-stimulated gene expression in intestinal tissue of autophagy-deficient mice in the absence of infection was mediated by the gut microbiota. Additionally, monocytes infiltrating into the autophagy-deficient intestinal microenvironment displayed an enhanced inflammatory profile and were necessary for protection against C. rodentium. Finally, we demonstrate that the microbiota-dependent IFN-I production that occurs in the autophagy-deficient host also protects against chemical injury of the intestine. Thus, autophagy proteins prevent a spontaneous IFN-I response to microbiota that is beneficial in the presence of infectious and non-infectious intestinal hazards. These results identify a role for autophagy proteins in controlling the magnitude of IFN-I signalling at the intestinal barrier.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ramanan, D. & Cadwell, K. Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe 19, 434–441 (2016).
Cadwell, K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat. Rev. Immunol. 16, 661–675 (2016).
Tal, M. C. et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl Acad. Sci. USA 106, 2770–2775 (2009).
Jounai, N. et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl Acad. Sci. USA 104, 14050–14055 (2007).
Liang, Q. et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15, 228–238 (2014).
Lei, Y. et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 36, 933–946 (2012).
Zhao, Y. et al. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 8, e1003086 (2012).
Mathew, R. et al. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol. Cell 55, 916–930 (2014).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Cadwell, K. et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010).
Kernbauer, E., Ding, Y. & Cadwell, K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98 (2014).
Marchiando, A. M. et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe 14, 216–224 (2013).
Grimm, W. A. et al. The Thr300Ala variant in ATG16L1 is associated with improved survival in human colorectal cancer and enhanced production of type I interferon. Gut 65, 456–464 (2016).
Lopez, C. A. et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253 (2016).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007).
Hoffmann, C. et al. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect. Immun. 77, 4668–4678 (2009).
Hwang, S. et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11, 397–409 (2012).
Choi, J. et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 40, 924–935 (2014).
Marino, G. et al. Autophagy is essential for mouse sense of balance. J. Clin. Invest. 120, 2331–2344 (2010).
Maurer, K. et al. Autophagy mediates tolerance to Staphylococcus aureus alpha-toxin. Cell Host Microbe 17, 429–440 (2015).
Cann, G. M. et al. Developmental expression of LC3alpha and beta: absence of fibronectin or autophagy phenotype in LC3beta knockout mice. Dev. Dyn. 237, 187–195 (2008).
Chen, Z. H. et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl Acad. Sci. USA 107, 18880–18885 (2010).
Inoue, J. et al. Autophagy in the intestinal epithelium regulates Citrobacter rodentium infection. Arch. Biochem. Biophys. 521, 95–101 (2012).
Pott, J., Kabat, A. M. & Maloy, K. J. Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in mice. Cell Host Microbe 23, 191–202 (2018).
Hubbard-Lucey, V. M. et al. Autophagy gene atg16l1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity 41, 579–591 (2014).
Matsuzawa-Ishimoto, Y. et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 214, 3687–3705 (2017).
Murthy, A. et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456–462 (2014).
Lassen, K. G. et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc. Natl Acad. Sci. USA 111, 7741–7746 (2014).
Zhang, H. et al. Myeloid ATG16L1 facilitates host–bacteria interactions in maintaining intestinal homeostasis. J. Immunol. 198, 2133–2146 (2017).
Gao, P. et al. The inflammatory bowel disease-associated autophagy gene Atg16L1T300A acts as a dominant negative variant in mice. J. Immunol. 198, 2457–2467 (2017).
Putt, K. S. et al. Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nat. Chem. Biol. 2, 543–550 (2006).
Konno, H., Konno, K. & Barber, G. N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 (2013).
Saitoh, T. et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA 106, 20842–20846 (2009).
Moretti, J. et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell 171, 809–823 (2017).
Prabakaran, T. et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 37, e97858 (2018).
Sun, L. et al. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe 17, 85–97 (2015).
Kamada, N. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012).
Kim, Y. G. et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity 34, 769–780 (2011).
Van den Bossche, J., O’Neill, L. A. & Menon, D. Macrophage immunometabolism: where are we (going)? Trends Immunol. 38, 395–406 (2017).
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008).
Liu, Z. et al. Role of inflammasomes in host defense against Citrobacter rodentium infection. J. Biol. Chem. 287, 16955–16964 (2012).
Lan, Y. Y., Londono, D., Bouley, R., Rooney, M. S. & Hacohen, N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 9, 180–192 (2014).
Zeng, M. Y. et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658 (2016).
Li, X. D. et al. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc. Natl Acad. Sci. USA 108, 17390–17395 (2011).
Steed, A. L. et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357, 498–502 (2017).
Samie, M. et al. Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat. Immunol. 19, 246–254 (2018).
Gentle, I. E. et al. TIR-domain-containing adapter-inducing interferon-beta (TRIF) forms filamentous structures, whose pro-apoptotic signalling is terminated by autophagy. FEBS J. 284, 1987–2003 (2017).
Yang, Q. et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 13, e1006600 (2017).
Benjamin, J. L., Sumpter, R. Jr, Levine, B. & Hooper, L. V. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe 13, 723–734 (2013).
Conway, K. L. et al. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology 145, 1347–1357 (2013).
Kimmey, J. M. et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569 (2015).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108, 4516–4522 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Vazquez-Baeza, Y., Pirrung, M., Gonzalez, A. & Knight, R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 16 (2013).
Acknowledgements
The authors acknowledge the following NYU facilities for use of their instruments and technical assistance: the Microscopy Core (RR023704), the Histopathology and Immunohistochemistry Core (P30CA016087, NIH S10 OD010584-01A1 and S10 OD018338-01), the Cytometry and Cell Sorting Laboratory (P30CA016087), the Genome Technology Center (P30CA016087) and the Gnotobiotic Facility (Colton Center for Autoimmunity). This work was supported by US National Institute of Health (NIH) grants R01 HL123340 (K.C.), R01 DK093668 (K.C.), R01 DK103788 (K.C.), R01 AI121244 (K.C.), F31 DK111139 (P.K.M.) and T32 AI100853 (E.R.), a Faculty Scholar grant from the Howard Hughes Medical Institute (K.C.), an Advanced Research Grant from the Merieux Institute (K.C.), a Rainin Foundation Innovator Award (K.C.), the Stony Wold-Herbert Fund (K.C.) and philanthropy from B. Levine (K.C.), and a Crohn’s & Colitis Foundation Research Fellowship Award (A.M.). K.C. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Author information
Authors and Affiliations
Contributions
P.K.M., A.M. and K.C. formulated the original hypothesis, designed the study and analysed the results. E.R. assisted with transcriptomics analyses. R.X. performed histopathology analyses. E.K. assisted with experiments involving GF mice. S.L.S. and F.Y. assisted with analyses of microbial communities. P.K.M. and K.C. wrote the manuscript, and all authors commented on the manuscript, data and conclusions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–6.
Supplementary Table 1
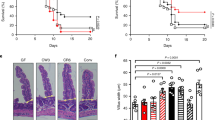
Gene expression dataset, related to Fig. 6. Shown are normalized log2 expression values for 812 altered genes from infiltrating inflammatory monocytes isolated from the colon of day 9 C. rodentium-infected WT and Atg16L1HM mice.
Supplementary Table 2
A dataset showing the exact P value for all graphs in this manuscript.
Rights and permissions
About this article
Cite this article
Martin, P.K., Marchiando, A., Xu, R. et al. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol 3, 1131–1141 (2018). https://doi.org/10.1038/s41564-018-0229-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0229-0
This article is cited by
-
The commensal skin microbiota triggers type I IFN–dependent innate repair responses in injured skin
Nature Immunology (2020)
-
Genome-wide association study of Buruli ulcer in rural Benin highlights role of two LncRNAs and the autophagy pathway
Communications Biology (2020)
-
Autophagy and microbial pathogenesis
Cell Death & Differentiation (2020)
-
High-throughput screening identified miR-7-2-3p and miR-29c-3p as metastasis suppressors in gallbladder carcinoma
Journal of Gastroenterology (2020)
-
Human genetics of Buruli ulcer
Human Genetics (2020)