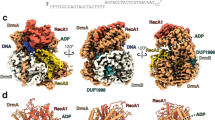
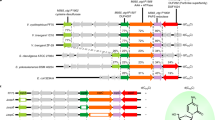
Abstract
The evolutionary pressure imposed by phage predation on bacteria and archaea has resulted in the development of effective anti-phage defence mechanisms, including restriction–modification and CRISPR–Cas systems. Here, we report on a new defence system, DISARM (defence island system associated with restriction–modification), which is widespread in bacteria and archaea. DISARM is composed of five genes, including a DNA methylase and four other genes annotated as a helicase domain, a phospholipase D (PLD) domain, a DUF1998 domain and a gene of unknown function. Engineering the Bacillus paralicheniformis 9945a DISARM system into Bacillus subtilis has rendered the engineered bacteria protected against phages from all three major families of tailed double-stranded DNA phages. Using a series of gene deletions, we show that four of the five genes are essential for DISARM-mediated defence, with the fifth (PLD) being redundant for defence against some of the phages. We further show that DISARM restricts incoming phage DNA and that the B. paralicheniformis DISARM methylase modifies host CCWGG motifs as a marker of self DNA akin to restriction–modification systems. Our results suggest that DISARM is a new type of multi-gene restriction–modification module, expanding the arsenal of defence systems known to be at the disposal of prokaryotes against their viruses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Labrie, S. J., Samson, J. E. & Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010).
Stern, A. & Sorek, R. The phage–host arms race: shaping the evolution of microbes. Bioessays 33, 43–51 (2011).
Dy, R. L., Richter, C., Salmond, G. P. C. & Fineran, P. C. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 1, 307–331 (2014).
Samson, J. E., Magadán, A. H., Sabri, M. & Moineau, S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687 (2013).
Tock, M. R. & Dryden, D. T. F. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472 (2005).
Dy, R. L., Przybilski, R., Semeijn, K., Salmond, G. P. C. & Fineran, P. C. A widespread bacteriophage abortive infection system functions through a type IV toxin–antitoxin mechanism. Nucleic Acids Res. 42, 4590–4605 (2014).
Sorek, R., Lawrence, C. M. & Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266 (2013).
Marraffini, L. A. CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015).
Swarts, D. C. et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261 (2014).
Goldfarb, T. et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183 (2015).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41, 4360–4377 (2013).
Roberts, R. J. et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31, 1805–1812 (2003).
Makarova, K. S., Wolf, Y. I., Snir, S. & Koonin, E. V. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193, 6039–6056 (2011).
Markowitz, V. M. et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122 (2012).
Caruthers, J. M. et al. Structure of the second domain of the Bacillus subtilis DEAD-box RNA helicase YxiN. Acta Crystallogr. F 62, 1191–1195 (2006).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Caruthers, J. M. & McKay, D. B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 (2002).
Selvy, P. E., Lavieri, R. R., Lindsley, C. W. & Brown, H. A. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 (2011).
Grazulis, S. et al. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl Acad. Sci. USA 102, 15797–15802 (2005).
Zaremba, M. et al. DNA cleavage by CgII and NgoAVII requires interaction between N- and R-proteins and extensive nucleotide hydrolysis. Nucleic Acids Res. 42, 13887–13896 (2014).
Roberts, R. J., Vincze, T., Posfai, J. & Macelis, D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2015).
Dürr, H., Flaus, A., Owen-Hughes, T. & Hopfner, K.-P. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 34, 4160–4167 (2006).
Weigel, C. & Seitz, H. Bacteriophage replication modules. FEMS Microbiol. Rev. 30, 321–381 (2006).
Jakutyte, L. et al. Bacteriophage infection in rod-shaped Gram-positive bacteria: evidence for a preferential polar route for phage SPP1 entry in Bacillus subtilis. J. Bacteriol. 193, 4893–4903 (2011).
Jakutytė, L. et al. First steps of bacteriophage SPP1 entry into Bacillus subtilis. Virology 422, 425–434 (2012).
Blow, M. J. et al. The epigenomic landscape of prokaryotes. PLoS Genet. 12, e1005854 (2016).
Noyer-Weidner, M., Jentsch, S., Kupsch, J., Bergbauer, M. & Trautner, T. A. DNA methyltransferase genes of Bacillus subtilis phages: structural relatedness and gene expression. Gene 35, 143–150 (1985).
Noyer-Weidner, M., Walter, J., Terschüren, P. A., Chai, S. & Trautner, T. A. M.phi 3TII: a new monospecific DNA (cytosine-C5) methyltransferase with pronounced amino acid sequence similarity to a family of adenine-N6-DNA-methyltransferases. Nucleic Acids Res. 22, 5517–5523 (1994).
Stewart, C. R. et al. The genome of Bacillus subtilis bacteriophage SPO1. J. Mol. Biol. 388, 48–70 (2009).
Thiaville, J. J. et al. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc. Natl Acad. Sci. USA 113, E1452–E1459 (2016).
Sathiamoorthy, S. & Shin, J. A. Boundaries of the origin of replication: creation of a pET-28a-derived vector with p15A copy control allowing compatible coexistence with pET vectors. PLoS ONE 7, e47259 (2012).
Dunlap, C. A., Kwon, S.-W., Rooney, A. P. & Kim, S.-J. Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. Int. J. Syst. Evol. Microbiol. 65, 3487–3492 (2015).
Itaya, M. Stable positional cloning of long continuous DNA in the Bacillus subtilis genome vector. J. Biochem. 134, 513–519 (2003).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014).
Fortier, L.-C. & Moineau, S. Phage production and maintenance of stocks, including expected stock lifetimes. Methods Mol. Biol. 501, 203–219 (2009).
Mazzocco, A., Waddell, T. E., Lingohr, E. & Johnson, R. P. Enumeration of bacteriophages using the small drop plaque assay system. Methods Mol. Biol. 501, 81–85 (2009).
Baym, M. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10, e0128036 (2015).
Miura, F., Enomoto, Y., Dairiki, R. & Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Tzipilevich, E., Habusha, M. & Ben-Yehuda, S. Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168, 186–199 (2017).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Acknowledgements
The authors thank M. Shamir, G. Amitai, S. Edelheit, A. Lopatina, T. Wein, Z. Erez and Z. Meir for discussions during the course of this study. The authors also thank A. Leavitt for discussions and assistance in adsorption assays and M. Keren for assistance with plaque assays. The authors thank I. Kolodkin-Gal for the pDR110 plasmid, J.M. Peters for the pJMP4 plasmid, S. Ben-Yehuda for the ET3 strain, P. Tavares for the lacO-containing SPP1 phage strain, T. Bucher, I. Pereman and E. Tzipilevich for their assistance with microscopy and O. Golani from the Weizmann Life Sciences Core Facilities for assistance in image analyses. This study was supported, in part, by the Israel Science Foundation (personal grants 1303/12 and 1360/16 and I-CORE grant 1796/12), the European Research Council (grant ERC-CoG 681203), the Minerva Foundation and by a research grant from the David and Fela Shapell Family Foundation.
Author information
Authors and Affiliations
Contributions
G.O. and S.M. designed the experiments, performed the experiments and analysed the results. H.S. identified, bioinformatically, the DISARM system. S.D. and G.O. performed bioinformatics analysis of DISARM systems. Z.M. performed bisulfite sequencing. S.S. assisted with experiments. G.Y. and G.O. performed microscopy experiments. G.O. and R.S. wrote the manuscript. R.S. supervised the study.
Corresponding author
Ethics declarations
Competing interests
R.S. is a scientific founder of BiomX and a member of its scientific advisory board.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Table 3, Supplementary Figures 1–10
Supplementary Table 1
Genes containing DUF1998, with no other domain, in microbial genomes
Supplementary Table 2
DISARM systems detected in microbial genomes
Rights and permissions
About this article
Cite this article
Ofir, G., Melamed, S., Sberro, H. et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol 3, 90–98 (2018). https://doi.org/10.1038/s41564-017-0051-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-017-0051-0
This article is cited by
-
Conservation and similarity of bacterial and eukaryotic innate immunity
Nature Reviews Microbiology (2024)
-
Discovery and structural mechanism of DNA endonucleases guided by RAGATH-18-derived RNAs
Cell Research (2024)
-
Engineered phage enzymes against drug-resistant pathogens: a review on advances and applications
Bioprocess and Biosystems Engineering (2024)
-
Challenges of bacteriophages application in controlling bacterial plant diseases and how to overcome them
Journal of Genetic Engineering and Biotechnology (2023)
-
Bacterial defences: mechanisms, evolution and antimicrobial resistance
Nature Reviews Microbiology (2023)