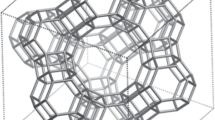
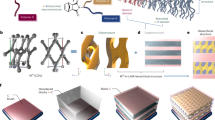
Abstract
The three-dimensional arrangement of natural and synthetic network materials determines their application range. Control over the real-time incorporation of each building block and functional group is desired to regulate the macroscopic properties of the material from the molecular level onwards. Here we report an approach combining kinetic Monte Carlo and molecular dynamics simulations that chemically and physically predicts the interactions between building blocks in time and in space for the entire formation process of three-dimensional networks. This framework takes into account variations in inter- and intramolecular chemical reactivity, diffusivity, segmental compositions, branch/network point locations and defects. From the kinetic and three-dimensional structural information gathered, we construct structure–property relationships based on molecular descriptors such as pore size or dangling chain distribution and differentiate ideal from non-ideal structural elements. We validate such relationships by synthesizing organosilica, epoxy–amine and Diels–Alder networks with tailored properties and functions, further demonstrating the broad applicability of the platform.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source data are provided with this paper. The remaining data that support the findings of this study are included within the manuscript and its supplementary files and available from the corresponding authors upon request.
Code availability
The software used to access the molecular configuration of 3D network polymers in this paper is freely accessible via https://lammps.sandia.gov. Representative input and processing scripts are available at https://github.com/ldkeer/NatMater_dekeer_2021.
References
Flory, P. J. Network topology and the theory of rubber elasticity. Br. Polym. J. 17, 96–102 (1985).
Kryven, I. Bond percolation in coloured and multiplex networks. Nat. Commun. 10, 404 (2019).
Zhong, M. J., Wang, R., Kawamoto, K., Olsen, B. D. & Johnson, J. A. Quantifying the impact of molecular defects on polymer network elasticity. Science 353, 1264–1268 (2016).
Sheiko, S. S. & Dobrynin, A. V. Architectural code for rubber elasticity: from supersoft to superfirm materials. Macromolecules 52, 7531–7546 (2019).
LLorca, J. On the quest for the strongest materials. Science 360, 264–265 (2018).
Berman, D., Erdemir, A. & Sumant, A. V. Graphene: a new emerging lubricant. Mater. Today 17, 31–42 (2014).
Anderson, M. W. & Klinowski, J. Direct observation of shape selectivity in zeolite ZSM-5 by magic-angle-spinning NMR. Nature 339, 200–203 (1989).
Rimer, J. D. Rational design of zeolite catalysts. Nat. Catal. 1, 488–489 (2018).
Boerjan, W., Ralph, J. & Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 (2003).
Burg, J. A. & Dauskardt, R. H. Elastic and thermal expansion asymmetry in dense molecular materials. Nat. Mater. 15, 974–980 (2016).
Owens, G. J. et al. Sol–gel based materials for biomedical applications. Prog. Mater. Sci. 77, 1–79 (2016).
Loccufier, E. et al. Silica nanofibrous membranes for the separation of heterogeneous azeotropes. Adv. Funct. Mater. 28, 1804138 (2018).
Caló, E. & Khutoryanskiy, V. V. Biomedical applications of hydrogels: a review of patents and commercial products. Eur. Polym. J. 65, 252–267 (2015).
D’hooge, D. R., Van Steenberge, P. H. M., Reyniers, M.-F. & Marin, G. B. The strength of multi-scale modeling to unveil the complexity of radical polymerization. Prog. Polym. Sci. 58, 59–89 (2016).
Matyjaszewski, K. Advanced materials by atom transfer radical polymerization. Adv. Mater. 30, 1706441 (2018).
Van Steenberge, P. H. M. et al. Visualization and design of the functional group distribution during statistical copolymerization. Nat. Commun. 10, 3641 (2019).
Di Lorenzo, F. & Seiffert, S. Nanostructural heterogeneity in polymer networks and gels. Polym. Chem. 6, 5515–5528 (2015).
Gu, Y., Zhao, J. & Johnson, J. A. Polymer networks: from rubbers and gels to porous frameworks. Angew. Chem. Int. Ed. 59, 5022–5049 (2020).
Sandakov, G. I., Smirnov, L. P., Sosikov, A. I., Summanen, K. T. & Volkova, N. N. NMR analysis of distribution of chain lengths between crosslinks of polymer networks. J. Polym. Sci. Polym. Phys. 32, 1585–1592 (1994).
Mathur, A. M. & Scranton, A. B. Characterization of hydrogels using nuclear magnetic resonance spectroscopy. Biomaterials 17, 547–557 (1996).
Tripathi, A. K., Neenan, M. L., Sundberg, D. C. & Tsavalas, J. G. Influence of n-alkyl ester groups on efficiency of crosslinking for methacrylate monomers copolymerized with EGDMA: experiments and Monte Carlo simulations of reaction kinetics and sol–gel structure. Polymer 96, 130–145 (2016).
Kryven, I., Duivenvoorden, J., Hermans, J. & Iedema, P. D. Random graph approach to multifunctional molecular networks. Macromol. Theor. Simul. 25, 449–465 (2016).
Van Steenberge, P. H. M. et al. Linear gradient quality of ATRP copolymers. Macromolecules 45, 8519–8531 (2012).
D’hooge, D. R. In silico tracking of individual species accelerating progress in macromolecular engineering and design. Macromol. Rapid Commun. 39, 1800057 (2018).
Fierens, S. K. et al. Model-based design to push the boundaries of sequence control. Macromolecules 49, 9336–9344 (2016).
Buback, M., Feldermann, A., Barner-Kowollik, C. & Lacik, I. Propagation rate coefficients of acrylate–methacrylate free-radical bulk copolymerizations. Macromolecules 34, 5439–5448 (2001).
Burg, J. A. et al. Hyperconnected molecular glass network architectures with exceptional elastic properties. Nat. Commun. 8, 1019 (2017).
Schwab, F. K. & Denniston, C. Reaction and characterisation of a two-stage thermoset using molecular dynamics. Polym. Chem. 10, 4413–4427 (2019).
Zanjani, M. B. et al. Computational investigation of the effect of network architecture on mechanical properties of dynamically cross-linked polymer materials. Macromol. Theor. Simul. 28, 1900008 (2019).
Oliver, M. S., Dubois, G., Sherwood, M., Gage, D. M. & Dauskardt, R. H. Molecular origins of the mechanical behavior of hybrid glasses. Adv. Funct. Mater. 20, 2884–2892 (2010).
Kilic, K. I. & Dauskardt, R. H. Design of ultrastiff organosilicate hybrid glasses. Adv. Funct. Mater. 29, 1904890 (2019).
Talati, M., Albaret, T. & Tanguy, A. Atomistic simulations of elastic and plastic properties in amorphous silicon. Europhys. Lett. 86, 66005 (2009).
Kawamoto, K., Zhong, M. J., Wang, R., Olsen, B. D. & Johnson, J. A. Loops versus branch functionality in model click hydrogels. Macromolecules 48, 8980–8988 (2015).
Dušek, K. & Patterson, D. Transition in swollen polymer networks induced by intramolecular condensation. J. Polym. Sci. Polym. Phys. 6, 1209–1216 (1968).
Panyukov, S. Loops in polymer networks. Macromolecules 52, 4145–4153 (2019).
Mita, I. & Horie, K. Diffusion-controlled reactions in polymer systems. J. Macromol. Sci. Polym. Rev. 27, 91–169 (1987).
Pouxviel, J. C., Boilot, J. P., Beloeil, J. C. & Lallemand, J. Y. NMR study of the sol/gel polymerization. J. Non-Cryst. Solids 89, 345–360 (1987).
Tang, J., Chen, Z., Fu, A. W. & Cheung, D. W. Capabilities of outlier detection schemes in large datasets, framework and methodologies. Knowl. Inf. Syst. 11, 45–84 (2007).
Xia, B. et al. Preparation of silica coatings with continuously adjustable refractive indices and wettability properties via sol–gel method. RSC Adv. 8, 6091–6098 (2018).
Yang, J., Lin, G.-S., Mou, C.-Y. & Tung, K.-L. Mesoporous silica thin membrane with tunable pore size for ultrahigh permeation and precise molecular separation. ACS Appl. Mater. Interfaces 12, 7459–7465 (2020).
Daelemans, L., Van Paepegem, W., D’hooge, D. R. & De Clerck, K. Excellent nanofiber adhesion for hybrid polymer materials with high toughness based on matrix interdiffusion during chemical conversion. Adv. Funct. Mater. 29, 1807434 (2019).
Li, J. & Mooney, D. J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 1, 16071 (2016).
Oehlenschlaeger, K. K. et al. Adaptable hetero Diels–Alder networks for fast self-healing under mild conditions. Adv. Mater. 26, 3561–3566 (2014).
Zhou, J. et al. Thermally reversible Diels–Alder-based polymerization: an experimental and theoretical assessment. Polym. Chem. 3, 628–639 (2012).
Zhou, J., Yu, G. & Huang, F. Supramolecular chemotherapy based on host–guest molecular recognition: a novel strategy in the battle against cancer with a bright future. Chem. Soc. Rev. 46, 7021–7053 (2017).
Harada, A., Takashima, Y. & Nakahata, M. Supramolecular polymeric materials via cyclodextrin–guest interactions. Acc. Chem. Res. 47, 2128–2140 (2014).
Chakma, P. & Konkolewicz, D. Dynamic covalent bonds in polymeric materials. Angew. Chem. Int. Ed. 58, 9682–9695 (2019).
Zou, W., Dong, J., Luo, Y., Zhao, Q. & Xie, T. Dynamic covalent polymer networks: from old chemistry to modern day innovations. Adv. Mater. 29, 1606100 (2017).
St John, N. A. & George, G. A. Cure kinetics and mechanisms of a tetraglycidyl-4,4′-diaminodiphenylmethane/diaminodiphenylsulphone epoxy resin using near i.r. spectroscopy. Polymer 33, 2679–2688 (1992).
Stockmayer, W. H. Theory of molecular size distribution and gel formation in branched-chain polymers. J. Chem. Phys. 11, 45–55 (1943).
Acknowledgements
L.D.K. acknowledges the research foundation Flanders (FWO; 1S37517N). P.H.M.V.S. and L.D. acknowledge FWO through a postdoctoral fellowship (12C4319N and 12ZR520N). D.R.D acknowledges FWO through G.0H52.16N and Flemish Government and Flanders Innovation & Entrepreneurship (Vlaio) through the Moonshot project P2C (HBC.2019.0114). The computational resources (Stevin Supercomputer Infrastructure) and services used in this work were provided by the Flemish Supercomputer Center, funded by Ghent University, FWO and the Flemish Government, department Economics, Science and Innovation (EWI). The work at Stanford University was supported by the US Department of Energy, Office of Basic Energy Sciences, under contract no. DE-FG02-07ER46391. C.B.-K. acknowledges an Australian Research Council Laureate Fellowship enabling his photochemical research programme as well as the Queensland University of Technology for key support. H.F. acknowledges Australian Research Council funding through a Discovery Early Career Researcher Award (DE200101096). We thank E. Loccufier for discussion regarding the interpretation of the experimental data for the membrane filtration. We thank J. Pelloth for carefully conducting initial kinetic experiments on the Diels–Alder chemistry.
Author information
Authors and Affiliations
Contributions
L.D.K, P.H.M.V.S and D.R.D. contributed to the development of matrix-based kMC simulations for network synthesis. L.D.K, P.H.M.V.S., M.-F.R. and D.R.D. focused on the determination of the scale-dependent model parameters of the kMC simulations and contributed to the construction of the associated structure–property relationships. D.R.D. developed the overall framework of the connection of kMC and MD simulations. K.I.K. and R.H.D. contributed to part of the MD simulations and interpretation, as well as the construction of the structure–property relationship for the organosilica case. L.D. and K.D.C. contributed to the experimental part concerning the epoxy–amine curing and the construction of the related structure–property relationship. D.K., H.F. and C.B.-K. conceptualized and conducted the experimental part concerning the Diels–Alder chemistry and the construction of the related structure–property relationships. All authors approved the manuscript and made revisions during its preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Materials thanks Simon Harrisson, Philippe Zinck and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–27, Discussion, Methods and Tables 1–5.
Source data
Source Data Fig. 3
Source data for Fig. 3.
Source Data Fig. 4
Source data for Fig. 4.
Source Data Fig. 5
Source data for Fig. 5.
Rights and permissions
About this article
Cite this article
De Keer, L., Kilic, K.I., Van Steenberge, P.H.M. et al. Computational prediction of the molecular configuration of three-dimensional network polymers. Nat. Mater. 20, 1422–1430 (2021). https://doi.org/10.1038/s41563-021-01040-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-021-01040-0
This article is cited by
-
Playing with Chlorine-Based Post-modification Strategies for Manufacturing Silica Nanofibrous Membranes Acting as Stable Hydrophobic Separation Barriers
Advanced Fiber Materials (2024)
-
Weaving tangled webs
Nature Materials (2022)