Abstract
Social distancing and isolation have been widely introduced to counter the COVID-19 pandemic. Adverse social, psychological and economic consequences of a complete or near-complete lockdown demand the development of more moderate contact-reduction policies. Adopting a social network approach, we evaluate the effectiveness of three distancing strategies designed to keep the curve flat and aid compliance in a post-lockdown world. These are: limiting interaction to a few repeated contacts akin to forming social bubbles; seeking similarity across contacts; and strengthening communities via triadic strategies. We simulate stochastic infection curves incorporating core elements from infection models, ideal-type social network models and statistical relational event models. We demonstrate that a strategic social network-based reduction of contact strongly enhances the effectiveness of social distancing measures while keeping risks lower. We provide scientific evidence for effective social distancing that can be applied in public health messaging and that can mitigate negative consequences of social isolation.
Similar content being viewed by others
Main
The non-pharmaceutical intervention of social distancing is a key policy to reduce the spread of COVID-19 by maintaining physical distance and reducing social interactions1. The aim is to slow transmission and the growth rate of infections to avoid overburdening healthcare systems—an approach widely known as flattening the curve2. Common social distancing measures are bans on public events, the closure of schools, universities and non-essential workplaces, limiting public transportation, travel and movement restrictions, and limiting physical interactions.
Social distancing interventions during previous outbreaks (for example, during SARS-CoV (severe acute respiratory syndrome coronavirus) in 2003) have often been based on expert recommendations rather than scientific evidence3. Existing research has mostly evaluated travel restrictions, school closures or vaccines4,5. Cancelling public gatherings and imposing travel restrictions decreases transmission and morbidity rates6, with mixed evidence on the efficacy of school closures7. Virtually no research exists on strategies based on individuals’ knowledge of their social surroundings, yet interventions are only effective when the public deems them acceptable8. Few have considered social networks, or if they did it was in relation to vaccinations9, contact tracing or analysing the spread of the virus8,10.
Since most facets of economic and social life require person-to-person contact, strategically reducing contacts is favourable to complete isolation. Increasing contact can likewise counter negative social, psychological and economic costs of quarantining individuals over prolonged periods of time and avoid compliance fatigue11. To achieve this aim, we propose behavioural network-based strategies for selective contact reduction that every individual and organization can easily understand, control and adopt. Applying insights from social and statistical network science, we demonstrate how changing network configurations of individuals’ contact choices and organizational routines can alter the rate and spread of the virus by providing guidelines to differentiate between high- and low-impact contacts for disease spread. We introduce and assess three strategies (contact with similar people; strengthening contact in communities; and repeatedly interacting with the same people in bubbles) that rely on less confinement and allow strategic social contact while still flattening the curve. Our approach balances public health concerns with social, psychological and economic needs for interpersonal interaction.
Flattening the (infection) curve operates to decrease the number of infected individuals at the height of the epidemic, by distributing the incidence of cases over a longer time horizon2. This is largely achieved by reducing the reproduction number (R), which represents how many individuals are infected by each carrier. Social distancing policies are implicitly designed to achieve this by limiting the amount of social contact between individuals. By introducing a social network approach, we propose that a decrease in R can simultaneously be achieved by managing the network structure of interpersonal contact.
From a social network perspective, the shape of the infection curve is closely related to the concept of network distance (or path lengths)12, which indicates the number of network steps needed to connect two nodes. Popularized examples of network distance include the six degrees of separation phenomenon13, which claims that any two people are connected through at most five acquaintances.
The relationship between infection curves and network distance can be illustrated with a simple network infection model (Fig. 1). Figure 1a,c depicts two networks with different path lengths, each with one hypothetically infected COVID-19 seed node (purple square). At each time step, the disease spreads from infected nodes to every node to which they are connected; thus, in the first step, the disease spreads from the seed node to its direct neighbours. In the second step, it spreads to their neighbours, who are at network distance 2 from the seed node, and so on. Over time, the virus moves along network ties until all nodes are infected. The example shows that the network distance of a node from the infection source (indicated by node colour in Fig. 1a,c) is identical to the number of time steps until the virus reaches it. The distribution of network distances to the source thus directly maps onto the curve of new infections (Fig. 1b,d).
a–d, Two example networks (a and c) have the same number of nodes (individuals) and ties (social interactions) but different structures (shorter path lengths in a and longer path lengths in c), which imply different infection curves (b and d, respectively). Bold ties highlight the shortest infection path from the infection source to the last infected individual in the respective networks. Network node colour indicates at which step a node is infected and maps onto the colours of the histogram bars.
In our example, both networks have the same number of nodes (individuals) and edges (interactions); however, the network depicted in Fig. 1c has a much flatter infection curve than the network depicted in Fig. 1a, even though all nodes are eventually infected in both cases. This is because the latter network has longer path lengths than the former one. In other words, there is more network distance between the individuals due to a differing structure of interaction, despite the same absolute contact prevalence. When adopting a network perspective, flattening the curve is thus equivalent to increasing the path length from an infected individual to all others, which can be achieved by restructuring contact (besides the general reduction of contact). Consequently, one aim of social distancing should be increasing the average network distance between individuals by smartly and strategically manipulating the structure of interactions. Our illustration shows a viable path to keep the COVID-19 curve flat while allowing some social interaction: we must devise interaction strategies that make real-life networks look more like the network in Fig. 1c, and less like the network in Fig. 1a.
We propose a series of strategies on how individuals can make local decisions to achieve this goal. Understanding which types of strategies of targeted contact reduction and social distancing are more efficient in increasing path lengths and flattening the curve can inform how to shift from short-term (complete lockdown) to long-term management of COVID-19 contagion processes. The contact reduction strategies we propose are based on insights into how items flow through networks, such as diseases, memes, information or ideas14,15,16,17. Such spread is generally hampered when networks consist of densely connected groups with few connections in-between (such as individuals who live in isolated villages scattered over sparse rural areas18). In contrast, contacts that bridge large distances are related to short paths and rapid spread. For instance, when commuters travel between these isolated villages, network distances decrease substantially14,18. Using this knowledge, we can avoid rapid contagion by encouraging social distancing strategies that increase clustering and reduce network shortcuts to reap the largest benefit of reducing social contact and limiting disease spread to a minimum. We propose three strategies aimed at increasing network clustering and eliminating shortcuts.
We outline the principles of the proposed strategies in Fig. 2. Figure 2a depicts a network in which densely connected communities are bridged by random, long-range ties. This type of network represents core features of real-world contact networks14 and is commonly known as a small-world network18. Within communities, individuals are similar to each other (as indicated by their node colour) and adjacent communities are geographically close (as indicated by node location). The further away two clusters are in the figure, the further they live from each other and the more dissimilar their members are. Figure 2a–d illustrates the successive, targeted contact reduction strategies, while the bar graph depicts the distribution of distances of all individuals from one of the two highlighted infection sources.
a–d, Based on an initial small-world network (a), example networks are mapped based on removing ties to dissimilar others who live far away (b), removing non-embedded ties that are not part of triads or four-cycles (c) or repeating rather than extending contact (d). Node colour represents an individual characteristic, where similarity in node colour represents similarity in this characteristic. Node placement represents geographic location of residence. Ties to dissimilar others who live far away are indicated by ties substantially longer than the average (that is, to nodes that are placed distantly and have very different colours). e, Bar graph showing network distances from the infection sources (highlighted in yellow in a–d) for the different scenarios.
In the first strategy (seek similarity; compare Fig. 2a and Fig. 2b), individuals choose their contact partners based on similarity of a predetermined individual characteristic19,20,21, such as those who live geographically close (spatial similarity), are members of the same organizations (for example, department at work) or are similar on continuous, highly variable demographic characteristics, such as age. Restructuring contact in this way reduces network bridges to groups of geographically distant others22 and to those with whom no organization or characteristic is shared; this contains the disease in localized areas of the network. A pre-requisite for this strategy is that people seek similarity on a dimension that facilitates forming many comparatively small groups (for example, based in neighbourhoods or small organizations). Segregation of large demographic groups, such as ethnic or racial segregation would not provide any measurable benefit. Further details are discussed in Box 1.
For the second strategy (strengthen communities; compare Fig. 2b and Fig. 2c), individuals must consider with whom their contact partners usually interact. When reducing contact, one should prioritize removing ties not embedded in triangles (a triangle is a network configuration of individuals i, j and h in which all three are mutually connected23,24). Thus, people should interact less with others who are not in contact with their other usual contact partners. For example, two friends should only meet if they have many other friends in common. Keeping contact in cohesive communities characterized by triangles can contain virus spread in local regions of the networks, rather than allowing it to spread to distant communities via network bridges25. This strategy is elaborated in Box 2.
For the third strategy (build bubbles through repeated contact; compare Fig. 2c and Fig. 2d), individuals must decide with whom they regularly want to interact and, over time, restrict interaction to those people. This reduces the number of contact partners rather than the number of interactions. This strategy of limiting contact to very few others with repeated interactions is in the spirit of a social contract with others to create social bubbles allowing only interactions within the same group delineated by common agreement. Similarly, employers could create contained departmental or work unit bubbles of employees. These micro-communities are difficult for a virus to penetrate and—importantly—if the infection is contracted by one contact, it is difficult for the virus to spread much further. Details of the strategy and comparisons with strategy 2 are presented in Box 3.
We now demonstrate how these three contact strategies impact infection curves using formal stochastic infection models that incorporate core elements from infection models, ideal-type network models and statistical relational event models. First, our model draws from classical disease modelling26,27 in which individuals (actors) can be in four states: susceptible; exposed (infected but not yet infectious); infectious; or recovered (no longer susceptible). At the start of the simulation, q actors are infectious while all others are susceptible. Susceptible actors can become exposed by having contact with infectious others; whether this contact results in contagion is determined probabilistically. A designated amount of time after becoming exposed, actors become infectious, and later move to the recovered state.
Second, as in many previous models of the dynamics of epidemics, contact probabilities in the population are imposed by a network structure that limits contact opportunities between actors28,29,30. This network represents the typical contact people had in a pre-COVID-19 world in different so-called social circles19,20,31. It consists of network ties between individuals who live geographically close, individuals who are similar on individual attributes, such as age, education or income, and individuals who are members of common groups, such as households and institutions (including schools and workplaces). Additionally, the network includes random connections in the population.
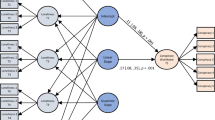
In the third component of the model, actors interact at discrete times with others from their personal network. During these meetings, the disease can be spread from infectious actors to susceptible alters. Notably, in contrast with other modelling approaches, actors do not interact with alters in their personal network with uniform probability (that is, at random). Rather, they are purposeful actors who make strategic choices about interaction partners. Choices are determined stochastically; strategies increase the likelihood of interacting with specific alters but are not deterministic. The mathematical formulation that determines contact choice follows earlier approaches used in network evolution32 and relational event models33,34. A flowchart of the model is presented in Fig. 4.
Squares indicate updating steps to individuals or the entire system. Diamond shapes represent decisions that determine the subsequent step in the simulation. In the iterative part of the model, a random individual i is chosen to initiate interactions with probability πcontact. When an interaction is initiated, a contact partner j is chosen with probability \(p\left( {i \to j} \right)\) following a multinomial choice model. If either interaction partner is infectious and the other is susceptible, contagion occurs with probability πinfection. Subsequently, among all individuals in the simulation, those who are in the exposed state for more than Texposure transition to the infectious state and those who are in the infectious state for more than Tinfection recover. These recursive steps are repeated until all individuals are either in the susceptible or recovered state. The colours red, green and yellow relate closely to the steps in the SEIR model, where red squares govern the transition from susceptible to exposed, the yellow square governs the transition from exposed to infectious, and the green square governs the transition from infectious to recovered. The purple square represents the step at which individuals strategically choose interaction partners to limit disease spread.
Our simulations explore the three interaction strategies we propose. First, in our seek similarity strategy, actors choose to interact predominantly with others who are similar to themselves based on one or several specified attributes. Second, actors can adopt our strengthen community strategy and choose to interact mostly with alters who have common connections in the underlying network. Third, adopting our repeat-contact bubble strategy, actors can base their choices on whom they have interacted with out of their previous contacts, both as senders and receives of interactions (see Methods). In our analyses, these three strategies are compared with a baseline case that mirrors a naive contact reduction strategy (in which individuals reduce interaction but choose randomly among their network contacts) and a null model that represents unbridled contact without any distancing. To make the interaction strategies comparable, we empirically calibrate statistical model parameters so that the average entropy in the probability distribution that represents the likelihood of different interaction choices is identical for all strategies (see Methods)35.
Following an initial analysis that represents a benchmark scenario of our disease model, we present a series of variations in modelling parameters that explore alternative scenarios and provide robustness checks. The benchmark scenario is conducted with 2,000 actors, and the variations and robustness analyses are conducted with 1,000 actors, unless otherwise specified.
Results
The average outcome of the benchmark scenario is presented in Fig. 4. The x axis represents time (as measured in simulation steps per actor) and the y axis shows the number of individuals infected at this time step out of a total population of 2,000. Curves are averaged over 40 simulation runs. The first scenario in blue shows a null or control interaction model in which there is no social distancing and actors interact at random. The other four strategies all employ a 50% contact reduction relative to the null model and compare different contact reduction strategies. The black line represents naive social distancing in which actors reduce contact in a random fashion. The golden line represents the infection curve when actors employ our first strategy (that is, seek similarity). The green line models our second triadic strategy of strengthening communities and represents the associated infection curve. Finally, the dark red line shows how infections develop when actors employ our third strategy of repeating contact in bubbles.
All three of our strategies substantially slow the spread of the virus compared with either no intervention or simple, non-strategic social distancing. The most effective approach is the strategic reduction of interaction with repeated contacts. Compared with the random contact reduction strategy, the average infection curve delays the peak of infections by 37%, decreases the height of the peak by 60% and results in 30% fewer infected individuals at the end of the simulation. This is marginally more efficient than the strengthening community strategy and the seeking similarity strategy, in this order (respective values: delay of peak: 34 and 18%, decrease in peak height: 49 and 44%; reduction of infected individuals: 19 and 2%). Note that these metrics cannot be interpreted as general estimates of the efficiency of these strategies in real-world networks.
Summarizing the sensitivity and robustness analyses presented below, strategic contact reduction has a substantive effect on flattening the curve compared with simple social distancing consistently across all scenarios. However, interesting variations occur. Full average infection curves and a description of the results for all model variations are presented in Extended Data Figs. 1–7 and Supplementary Information.
Different operationalizations of homophily
In the benchmark model, the seek similarity strategy was employed on one demographic attribute. However, in real-world social networks, individuals are homophilous on multiple characteristics36. Furthermore, the benchmark model only uses demographic homophily, while we previously also discussed the importance of geographic homophily. In a variation of the seek similarity strategy, we show that using geographic homophily for contact reduction is highly efficient—much more so than homophily based on demographic attributes (Extended Data Fig. 1b). Geographic homophily or similarity effectively eliminates contacts with distant others in the network. In a further analysis, we compare the benefits of using one dimension of demographic homophily or a composite of two dimensions that structure the network. This explores whether we should focus on interacting with persons similar in one dedicated dimension or seek out others who are similar in multiple dimensions simultaneously. Encouragingly, the focus on one strategic dimension of homophily provides similar outcomes to reducing demographic distance on both dimensions. In our limited example, this means that homophily can be encouraged only on the dimension that has lesser adverse consequences for societal cohesion, as opposed to reduction on both dimensions. Infection curves are presented in Extended Data Fig. 1c,d.
Employing mixed strategies
Since most individuals in a post-lockdown world need to interact across multiple social circles (for example, workplace, extended family and so on), employing only one strategy might not be practical. A mix of different strategies could therefore be more realistic for everyday use. We tested how four possible combinations of mixing strategies (three two-way combinations and one three-way combination) compare with the single strategies of seeking similarity and strengthening communities. We found that the combined strategies are comparably as effective as single strategies (see Extended Data Fig. 2) and can be recommended as alternatives if single strategies are not practicable in some contexts. Importantly, each combination performs better in limiting infection spread compared with the naive contact reduction strategy.
Varying the number of actors in the simulation
The computational complexity of our simulation prohibits assessing disease dynamics in very large networks (for example, 100,000+ actors), even on large distributed systems. Nevertheless, we can compare simulations using the same local network topology as the benchmark model on networks of 500, 1,000, 2,000 and 4,000 actors. Reassuringly, we find no variation of the relative effectiveness of the different interaction strategies by network size (see Extended Data Fig. 3). While this does not fully allow extrapolation to very large networks, it provides initial support that disease spread under the model could be similar within differently sized sub-regions of larger, real-world networks.
Varying the underlying network structure
The generation process of the ideal-type network that provides the opportunity structure among individuals with whom they can interact contains multiple degrees of freedom. These include the average number of contacts and the importance of different foci (geography, groups and attributes) in structuring contact. We provide infection curves for multiple scenarios in Extended Data Figs. 4 and 5, showing that our strategies work largely independent of the underlying structure. A first noteworthy finding from these simulations is that in networks with fewer connection opportunities, all strategies have much larger benefits compared with networks with more connection opportunities (Extended Data Fig. 4c,d). In fact, the strengthening community strategy does not seem to work anymore in scenarios with very high average connectivity in the underlying network—probably because of a large number of closed triangles. This shows that in communities that have lower connectivity, spread can be contained even more effectively. As a second finding, we see that in cases where the underlying network is not structured by homophily, the seeking similarity strategy does not work (Extended Data Fig. 5c), illustrating how the strategy relies on predetermined structural network features.
Variation in infectiousness and the length of the exposed period
Differences in infectiousness of the virus, and variations of the time during which individuals are in the exposed state relative to the infectious state do not influence the relative effectiveness of the different strategies, and average infection curves are presented in Extended Data Figs. 6 and 7, respectively.
Discussion
In the absence of a vaccine against COVID-19, governments and organizations face economic and social pressures to gradually and safely open up societies, yet they lack scientific evidence on how to do this. We provide clear social network-based strategies to empower individuals and organizations to adopt safer contact patterns across multiple domains by enabling individuals to differentiate between high- and low-impact contacts. The result may also be higher compliance since it empowers individuals to strategically adjust and control their own interactions without being requested to fully isolate. Instead of blanket self-isolation policies, the emphasis on similar, community-based and repetitive contacts is easy to understand and implement, thus making distancing measures more palatable over longer periods of time.
How can this be applied to real-world settings? When a firm lockdown is no longer mandated or recommended, individuals will want or need to interact in different social circles (for example, at the workplace or with wider family). In some of these settings, seeking similarity might not be possible (for example, in schools in which teachers and students of different ages come together). Consequently, the simple one-at-a-time strategic recommendations we analysed in most simulations might be impossible to strictly follow for some. Our sensitivity analysis using mixed strategies addresses this concern. For example, does mixing the three strategies still provide benefits or do they counteract one another? Reassuringly, our results show that a mix of strategies still provide comparable benefits to single strategies, and all work considerably better than simply releasing a floodgate of full non-strategic contact; however, further modelling is needed to assess the implications across a variety of contexts. When approaching this issue from a policy perspective, the design of steps to ease lockdown can be done with potential behavioural recommendations in mind: if network structures and demographic characteristics of individuals in particular regions suggest that the use of one strategy will yield the best results, decisions on which contact opportunities to allow (such as opening schools or local shops) might be taken so that this strategy can be adhered to most easily.
A second discussion point concerns potential unintended consequences of recommending our strengthening community and seeking similarity strategies. Our analyses and reasoning clearly should not be used to justify any form of racial or social group segregation or similar vulgar ideas. Beyond the obvious ethical and social consequences, segregation into such large groups would not be effective in curbing the spread of the virus, since strategic contact reduction relies on limiting contact to many small connected network regions not splitting into large groups. We acknowledge that advocating the creation of small communities and contact with mostly similar others on some dimensions could potentially result in the long-term reduction of intergroup contact and an associated rise in inequality37. In our simulations, we explored this concern by comparing the scenarios when homophilous ties in the underlying network are formed following similarity in multiple dimensions (for example, age and income). Our test of whether minimizing the overall difference in the two modelled attributes of contacts versus only reducing homophily on one dimension suggests that choosing one salient attribute can already be very effective. These findings provide preliminary evidence that policymakers could make smart choices relevant to their local context in deciding which attribute people should pay attention to, keeping the potential social consequences in mind. Nevertheless, combining similarity on two simulated individual-level attributes into a single indicator is still very likely to understate the complexity of how multiple individual traits intersect, and structure social interaction. Our conclusions about the intersectionality of multiple individual traits for disease spread remain tentative. This highlights that understanding the long-term social consequences of which types of public spaces are opened and, accordingly, which types of interaction are allowed requires more research and should be a chief concern in policy-making. Taking all of these considerations into consideration, for the moment, our simulations that explore the effect of increasing geographic proximity and the theoretical appeal of seeking similarity on residential location would make geographic similarity the preferred dimension when giving guidance to policymakers.
Third, a shortcoming of our simulation study is the limited number of network actors. While we varied the number of nodes from 500 to 4,000 and found no substantial difference in the results, we do not know the dynamics of the model in large networks of, for example, 100,000+ actors. In the current implementation of the model, the computational complexity increases more than linearly with the number of actors, which makes simulations with such numbers unrealistic. Consequently, algorithmic work on the model implementation is needed to extend its applicability to large, real-world networks, offering clear extensions for future research.
Despite these limitations, some concrete policy guidelines can be deduced from our network-based strategies. For hospital or essential workers, risk can be minimized by introducing shifts with a similar composition of employees (that is, repeating contact and creating bubbles) and distributing people into shifts based on, for example, residential proximity where possible (that is, seeking similarity). In workplaces and schools, staggering shifts and lessons with different start, end and break times by discrete organizational units and classrooms will keep contact in small groups and reduce contact between them. When providing private or home care to the elderly or vulnerable, the same person should visit rather than rotating or taking turns, and that person should be the one with fewest bridging ties to other groups and who lives the closest (geographically). Repeated social meetings of individuals of similar ages who live alone carry a comparatively low risk. However, in a household of five, when each person interacts with disparate sets of friends, many shortcuts are being formed that are potentially connected to a very high risk of spreading the disease.
In summary, simple behavioural rules can go a long way in keeping the curve flat. As the pressure increases throughout a pandemic to ease stringent lockdown measures, to relieve social, psychological and economic burdens, our approach provides insights to individuals, governments and organizations about three simple strategies: seeking similarity; strengthening interactions within communities; and repeated interaction with the same people to create bubbles.
Methods
Generation of stylized networks
The stylized binary network x that represents interaction opportunities is based on the typical contact people had in a pre-COVID-19 world. It is generated stochastically as the composite of four sub-processes that follow fairly standard ideal-type network-generating approaches. Representing place of residence, actors are assumed to have a fixed geographic location, as determined by coordinates in a two-dimensional space. They are members of groups (such as households) and institutions (such as schools or workplaces) and have individual attributes (such as age, education or income). Network ties are generated so that actors have some connections to geographically close alters, some ties to members of the same groups (representing, for example, co-workers), some ties to alters with similar attributes (for example, similar age) and some ties to random alters in the population. Jointly, these sub-processes create networks that have realistic values of local clustering, path lengths and homophily. All ties in the network are defined as undirected. The number of actors in the network is denoted by n. For the benchmark scenario presented in Fig. 4, n = 2,000, and for the variations and robustness analyses, n = 1,000, unless otherwise stated. In particular, the network sub-processes are defined as follows.
The first sub-process represents tie formation based on geographic proximity38. First, all actors in the network are randomly placed into a two-dimensional square. Second, each actor draws the number of contacts it forms in this sub-process dgeo,i from a uniform distribution between dgeo,min and dgeo,max; for example, if dgeo,min = 10 and dgeo,max = 20, every actor forms a random number of ties between 10 and 20 in this sub-process. Third, the user-defined density in geographic tie-formation dgeo defines the geographic proximity of contacts drawn, so that actor i randomly forms dgeo,i ties among those dgeo,i/dgeo that are close in Euclidean distance from actor i. For example, if actor i is posed to form dgeo,i = 12 ties and dgeo = 0.5, the actor randomly choses 12 out of the 24 closest alters to form a tie to. Across all simulated networks, we set dgeo = 0.3. Fourth, unilateral choices (where only i selected j but not vice versa) are symmetrized so that a non-directed connection exists between the actors.
The second sub-process represents tie formation in organizational foci (for example, workplaces)39. First, each actor is randomly assigned to a group so that all groups have on average m members. Second, each actor forms ties at random to other members within the same groups with a probability of ggroups. For example, when m = 10 and ggroups = 0.5, a tie from each actor to every alter in the same group is formed with a probability of 50%. Third, unilateral ties are symmetrized as above.
The third sub-process represents tie formation based on homophily (that is, seeking similarity); for example, similarity in age or income21. First, each actor is assigned an individual attribute ai between 0 and 100 with uniform probability (the scale of ai cancels later in the model). Second, for each actor, the normalized similarity simi,j to all alters j is calculated, which is 1 minus the absolute difference between ai and aj for actor j, divided by 100 (the range of the variable), so that simi,j = 1 when i and j have the identical value of a, and simi,j = 0 if they are at opposite ends of the scale. Third, each actor draws the number of contacts it forms in this sub-process dhomo,i from a uniform distribution between dhomo,min and dhomo,max. Fourth, each actor creates dhomo,i ties to alters j in the networks with a probability that is proportional to (simij)w, where higher values of w mean that individuals prefer more similar others. Across all reported simulations, we set w = 2. Fifth, unilateral ties are symmetrized as above.
The fourth sub-process represents haphazard ties that are not captured by any of the above processes. Here, simply, z ties per actor are created with respect to randomly chosen alters.
Definition of simulation model
Let the binary network x represent interaction opportunities between n individuals, labelled from 1 to n. Each node i can be characterized by a set of attributes \(\left( {a_i^k} \right)\) (for example, age or location).
Our model aims to reproduce the process of individuals interacting with some of these potential contacts. Similar to the classic SIR model26 (in which individuals are susceptible, infectious or recovered) and its SEIR extension27 (in which they are susceptible, exposed, infectious and then recovered), we assume that individuals can be in four different states: either susceptible to the disease, exposed (infected but not yet infectious), infectious or recovered. Infection occurs through social interactions, which are modelled in a similar fashion to the dynamic actor-oriented model34 developed for relational events. More specifically, our model comprises the following steps:
- 1.
At each step of the process, one individual is picked at random and initiates an interaction with the probability πcontact.
- 2.
An actor initiating an interaction can only pick one interaction partner. Only potential partners as defined by the network x can be chosen. The decision to interact is unilateral and depends on characteristics of the two persons through a probability model p.
- 3.
An infectious individual infects a healthy person when they interact, who then becomes exposed. This contagion occurs with the probability πinfection.
- 4.
After a fixed number of steps Texposure, an exposed individual becomes infectious.
- 5.
After becoming infectious, recovery occurs within Trecovery steps. Once recovered, individuals can no longer be infected.
- 6.
The process ends once there is no longer anyone exposed or infectious.
The steps of the model are illustrated in Fig. 3. Note that the mechanics of the infection align with previously proposed agent-based versions of the SIR and SEIR models40,41. Together, the probabilities πcontact and πinfection play a similar role to the classic infectivity rate β in SIR and SEIR models. The rate β models the average number of contacts per person (modelled here through πcontact) and the likelihood of infection (represented by πinfection); however, the equivalence is not direct due to the added step of the interaction probability p. The exposure and recovery times replace the classic exposure and recovery rates (often traditionally denoted as σ and γ) in a straightforward manner.
We turn to the definition of the probability model p. Let Ni be the set of potential contacts, or alters j of a given individual i in the network x. We define for each step t of the process: Li(j,t) as the number of previous interactions between i and an alter j, within the past λ interactions of i. In our simulations, the number λ was arbitrarily set to 2 but can be adjusted easily in the replication files.
For each alter \(j \in N_i\), the value s(i,j) represents the statistic driving the strategical choice of i to pick j. Specifically, we define three different ways depending on whether the homophily, triadic (that is, strengthening community) or repetition bubble strategy is chosen (however, other arbitrary statistics can be defined). The statistic ssimilarity accounts for the level of similarity between i and j given a set of attributes; scommunity corresponds to the number of alters they share, and srepetition is the count of previous interactions within the past λ contacts of i. In practice, these statistics are calculated as:
The probability for i to pick j is defined as a multinomial choice probability42, follo wing the logic of previous relational event34 and stochastic network models32. The intuition behind this distribution is that each potential partner in Ni is assigned an objective function value, and choosing a partner is based on these values. Mathematically, the objective function is an exponentiated linear function of the statistic s(i,j), weighted by a parameter α. We further assume that individuals can reduce a certain percentage of their interactions. Considering the probability πcontact of initiating an interaction in the first place, the relevant probability distribution becomes:
These probabilities can be loosely interpreted in terms of log-transformed odds ratios, similar to logit models. Given two potential partners j1 and j2 for whom the statistic s increases by one unit (that is, s(i,j2) = s(i,j1) + 1), the following log ratio simplifies to:
For example, if we use s = srepetition and αrepetition = log[2], the probability of picking one alter present in the past contacts of i is twice as high as picking another alter who is not.
Calibration of model parameters
The strategy of picking an interaction partner at random corresponds to the model without any statistic s, reducing the probability distribution to a uniform 1. For the three other strategies, the parameters αsimilarity, αcommunity and αrepetition are adjusted to keep the models comparable.
To this end, we use the measure of explained variation for dynamic network models devised by Snijders35. This measure builds on the Shannon entropy and can be applied to our model to assess the degree of certainty in individual’s choices. For a given individual i at a step t, this measure is defined as:
Intuitively, this measure equals 0 in the case of the random strategy where the probability of picking any alter is identical. It increases whenever some outcomes are favoured over others and equals 1 if one outcome has all of the probability mass.
Since the model assumes that all individuals are equally likely to initiate interactions, we can average this measure over all actors. Moreover, in the case of the repetition strategy, the measure is time dependent. Thus, we use its expected value over the whole process. We finally use the following aggregated measure to evaluate the certainty of outcomes of a specific strategy:
For this article, we first fix the parameter αrepetition at a value of 2.5 and calculate an estimated value \(\widehat {R_{\rm{H}}}\left( {\pi _{{\rm{contact}}},\alpha _{{\rm{repetition}}}} \right)\) of this measure. This experience-based parameter choice results in an associated RH value between 0.3 and 0.5 in the different scenario, which is realistic in terms of size (see the definition above). To compare this model with others, we then define the parameters αsimilarity and αcommunity that verify:
using a standard optimization algorithm. The average parameters across simulations for the different network scenarios are αcommunity = 0.75 and αsimilarity = 17.6. While the latter parameter appears large, note that the associated statistic ssimilarity ranges from 0 to 1, with most realized values close to 1. The R code associated with all of the calculations is provided in the online repository referenced in the Code Availability statement.
Parametrization of the different simulations
Unless otherwise specified, all simulations use πcontact = 0.5 except for the null model, which uses πcontact = 1. In all simulations except those that vary the infectiousness, πinfection = 0.8. Unless otherwise noted, Texposure = 1n and πinfection = 4n. Given the substantial computational burden involved in conducting the simulations, 48 repetitions were run for networks with n ≤ 1,000, with 40 for larger networks. Experiments varying Texposure and πinfection used 24 repetitions.
For the experiments that vary the structure of the underlying network and the network size, the parameters that guide the stochastic network creation are presented in Supplementary Table 1. Descriptive statistics of these networks are presented in Supplementary Table 2. The underlying networks that are used in the other variation experiments are generated according to the parameters denoted ‘1: baseline’ in Supplementary Table 1.
The four experiments that vary the time during which individuals are in the exposed state before becoming infectious use values for Texposure of 0, 1n, 2n, 3n and 4n.
The four experiments that vary the infectiousness of the disease use values for πinfection of 0.55, 0.65, 0.80 and 0.95.
The experiment that used geography as the basis of the homophily strategy was created according to the ‘1: baseline’ parameters but used the Euclidean distance in geographic placement as the basis for choosing interaction partners in the homophily strategy. The two experiments on multidimensional homophily used underlying networks created following the ‘1: baseline’ parameters, with the exception that instead of one homophilous attribute, two attributes were defined and the number of ties created according to the homophily parameter was split evenly between the two dimensions. The homophily strategy used for the simulated infection curves in the two scenarios differs in the sense that in the first, individuals interact according to minimizing the absolute difference in both attributes. In the second scenario, only the first attribute is used as the basis of the homophily strategy and the second attribute is ignored.
For the experiments using mixed strategies, the probability of partner choice \(p\left( {i \to j} \right)\) can depend on a vector of statistics and parameters34. The entropy based on a set parameter vector was used to calibrate the parameter for the homophily and triadic closure strategy as comparison cases. Parameter choices rely on experimentation to result in similar entropy values to when using single strategies. For the mixed strategy of repetition and homophily, the parameters were set to αsimiliarity = 7 and αrepetition = 1.6. For the mixed strategy of repetition and triadic closure, the parameters were set to αcommunity = 0.35 and αrepetition = 1.6. For the mixed strategy of homophily and triadic closure, the parameters were set to αsimiliarity = 6 and αcommunity = 0.35. For the mixed strategy incorporating all three, the parameters were set to αsimiliarity = 4, αcommunity = 0.3 and αrepetition = 1.2.
The simulated average infection curves for all experiments can be found in Extended Data Figs. 1–7. Descriptive results for the simulations, in terms of delay of peak, height of peak and total number infected at the end of the simulation, are presented in Supplementary Table 3. Note that the descriptive statistics in this table present the averages of characteristics of the repetitions of the simulated infection curves, which are not the same as the characteristics of the average infection curves as presented in Extended Data Figs. 1–7.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
No empirical data was used in this article, only simulated data. Routines to produce the simulated data are fully disclosed in the online resource presented in the Code Availability section.
Code availability
The replication files for this paper, including the R code associated with all calculations and customized functions in the statistics environment R and an example script, are available on Zenodo (https://zenodo.org/record/3782465), a general-purpose open-access repository developed under the European OpenAIRE programme and operated by CERN.
References
Glass, R. J., Glass, L. M., Beyeler, W. E. & Min, H. J. Targeted social distancing design for pandemic influenza. Emerg. Infect. Dis. 12, 1671–1681 (2006).
Roberts, S. Flattening the coronavirus curve. The New York Times https://www.nytimes.com/article/flatten-curve-coronavirus.html (27 March 2020).
World Health Organization Writing Group et al. Non-pharmaceutical interventions for pandemic influenza, national and community measures. Emerg. Infect. Dis. 12, 88–94 (2012).
Ferguson, N. M. et al. Strategies for mitigating an influenza pandemic. Nature 442, 448–452 (2006).
Germann, T. C., Kadau, K., Longini, I. M. & Macken, C. A. Mitigation strategies for pandemic influenza in the United States. Proc. Natl Acad. Sci. USA 103, 5935–5940 (2006).
Aledort, J. E., Lurie, N., Wasserman, J. & Bozzette, S. A. Non-pharmaceutical public health interventions for pandemic influenza: an evaluation of the evidence base. BMC Public Health 7, 208 (2007).
Jackson, C., Vynnycky, E., Hawker, J., Olowokure, B. & Mangtani, P. School closures and influenza: systematic review of epidemiological studies. BMJ Open 3, e002149 (2013).
Sun, K., Chen, J. & Viboud, C. Early epidemiological analysis of the coronavirus disease 2019 outbreak based on crowdsourced data: a population-level observational study. Lancet Digital Health 2, e201–e208 (2020).
Ventresca, M. & Aleman, D. Evaluation of strategies to mitigate contagion spread using social network characteristics. Soc. Netw. 35, 75–88 (2013).
Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. J. Am. Med. Assoc. 323, 1239–1242 (2020).
Morse, S. S., Garwin, R. L. & Olsiewski, P. J. Public health. Next flu pandemic: what to do until the vaccine arrives? Science 314, 929 (2006).
Wasserman, S. & Faust, K. Social Network Analysis: Methods and Applications (Cambridge Univ. Press, 1994).
Milgram, S. The small world problem. Psychol. Today 2, 60–67 (1967).
Watts, D. J. & Strogatz, S. H. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 (1998).
Podolny, J. M. Networks as the pipes and prisms of the market. Am. J. Sociol. 107, 33–60 (2001).
Borgatti, S. P. Centrality and network flow. Soc. Netw. 27, 55–71 (2005).
Centola, D. The spread of behavior in an online social network experiment. Science 329, 1194–1197 (2010).
Watts, D. J. Networks, dynamics, and the small-world phenomenon. Am. J. Sociol. 105, 493–527 (1999).
Feld, S. The focused organization of social ties. Am. J. Sociol. 86, 1015–1035 (1981).
Rivera, M. T., Soderstrom, S. B. & Uzzi, B. Dynamics of dyads in social networks: assortative, relational, and proximity mechanisms. Annu. Rev. Sociol. 36, 91–115 (2010).
McPherson, M., Smith-Lovin, L. & Cook, J. M. Birds of a feather: homophily in social networks. Annu. Rev. Sociol. 27, 415–444 (2001).
Pellis, L., Cauchemez, S., Ferguson, N. M. & Fraser, C. Systematic selection between age and household structure for models aimed at emerging epidemic predictions. Nat. Commun. 11, 906 (2020).
Granovetter, M. S. The strength of weak ties. Am. J. Sociol. 78, 1360–1380 (1973).
Goodreau, S. M., Kitts, J. A. & Morris, M. Birds of a feather, or friend of a friend? Using exponential random graph models to investigate adolescent social networks. Demography 46, 103–125 (2009).
Burt, R. S. Structural Holes: The Social Structure of Competition (Harvard Univ. Press, 1992).
Kermack, W. O., McKendrick, A. G. & Walker, G. T. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. A 115, 700–721 (1927).
Anderson, R. M. & May, R. M. Infectious Diseases of Humans: Dynamics and Control (Oxford Univ. Press, 1992).
Newman, M. E. J. Spread of epidemic disease on networks. Phys. Rev. E 66, 016128 (2002).
Halloran, M. E. et al. Modeling targeted layered containment of an influenza pandemic in the United States. Proc. Natl Acad. Sci. USA 105, 4639–4644 (2008).
Salathé, M. et al. A high-resolution human contact network for infectious disease transmission. Proc. Natl Acad. Sci. USA 107, 22020–22025 (2010).
Block, P. Network evolution and social situations. Sociol. Sci. 5, 402–431 (2018).
Snijders, T. A. B. The statistical evaluation of social network dynamics. Sociol. Methodol. 31, 361–395 (2001).
Butts, C. T. A relational event framework for social action. Sociol. Methodol. 38, 155–200 (2008).
Stadtfeld, C. & Block, P. Interactions, actors, and time: dynamic network actor models for relational events. Sociol. Sci. 4, 318–352 (2017).
Snijders, T. A. B. Explained variation in dynamic network models. Math. Sci. Hum. 42, 5–15 (2004).
Block, P. & Grund, T. Multidimensional homophily in friendship networks. Net. Sci. 2, 189–212 (2014).
DiMaggio, P. & Garip, F. Network effects and social inequality. Annu. Rev. Sociol. 38, 93–118 (2012).
Hamill, L. & Gilbert, N. Social circles: a simple structure for agent-based social network models. J. Artif. Soc. Soc. Simul. 12, 23 (2009).
Hébert-Dufresne, L. & Althouse, B. M. Complex dynamics of synergistic coinfections on realistically clustered networks. Proc. Natl Acad. Sci. USA 112, 10551–10556 (2015).
Chowell, G., Sattenspiel, L., Bansal, S. & Viboud, C. Mathematical models to characterize early epidemic growth: a review. Phys. Life Rev. 18, 66–97 (2016).
Siettos, C. I. & Russo, L. Mathematical modeling of infectious disease dynamics. Virulence 4, 295–306 (2013).
McFadden, D. L. in Frontiers in Econometrics 105–142 (Academic Press, 1974).
Acknowledgements
P.B., J.B.D., C.R., R.K. and M.C.M. are supported by Leverhulme Centre for Demographic Science (The Leverhulme Trust). C.R. is supported by the British Academy. M.C.M. is supported by ERC Advanced Grant CHRONO (835079). We thank M. Verhagen, V. Rotondi and L. Andriano for feedback. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
P.B., M.H. and I.J.R. conceptualized the study. P.B. and M.H. contributed methodology and implementation and performed analyses. P.B., M.H., I.J.R. and M.C.M. wrote the initial manuscript and provided visualization. All authors (P.B., M.H., I.J.R., J.B.D., C.R., R.K. and M.C.M.) discussed the research design and reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Peer review reports are available. Primary Handling Editor: Charlotte Payne.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Average infection curves varying by the operationalisation of homophily.
Curves compare 4 contact reduction strategies to the null model of no social distancing, as described in the main text. (a) Reference model with standard operationalisation of homophily; (b) model with homophily based on geographic proximity; (c) underlying network model with homophily based on two dimensions, interaction strategy minimises the overall difference along both attributes; (d) underlying network model with homophily based on two dimensions, interaction strategy minimises the difference only on the first attribute.
Extended Data Fig. 2 Average infection curves when employing mixed strategies.
Curves compare 4 contact reduction strategies to the null model of no social distancing, as described in the main text. (a) Mixed strategy of repetition and triadic closure; (b) mixed strategy of repetition and homophily; (c) mixed strategy of repetition, homophily, and triadic closure; (d) mixed strategy of homophily and triadic closure.
Extended Data Fig. 3 Average infection curves of networks of different size.
Curves compare 4 contact reduction strategies to the null model of no social distancing, as described in the main text. (a) 500 actors; (b) 1000 actors; (c) 2000 actors; (d) 4000 actors.
Extended Data Fig. 4 Average infection curves of networks with different topologies.
Curves compare 4 contact reduction strategies to the null model of no social distancing, as described in the main text. Names refer to the parametrisation given in Supplementary Table 1. (a) baseline scenario; (b) random network; (c) higher degree; (d) lower degree.
Extended Data Fig. 5 Average infection curves of networks with different topologies.
Curves compare 4 contact reduction strategies to the null model of no social distancing, as described in the main text. Names refer to the parametrisation given in Supplementary Table 1. (a) no groups; (b) no geography; (c) small world-ish.
Extended Data Fig. 6 Average infection curves of networks with varying levels of infectiousness of the virus.
Curves compare 4 contact reduction strategies to the null model of no social distancing, as described in the main text. (a) πinfection = 0.55; (b) πinfection = 0.65; (c) πinfection = 0.8; (d) πinfection = 0.95.
Extended Data Fig. 7 Average infection curves of networks with varying levels of time actors spend in state “exposed”.
Curves compare 4 contact reduction strategies to the null model of no social distancing, as described in the main text. (a) Texposed = 0; (b) Texposed = 1n; (c) Texposed = 2n; (d) Texposed = 3n; (e) Texposed = 4n.
Supplementary information
Rights and permissions
About this article
Cite this article
Block, P., Hoffman, M., Raabe, I.J. et al. Social network-based distancing strategies to flatten the COVID-19 curve in a post-lockdown world. Nat Hum Behav 4, 588–596 (2020). https://doi.org/10.1038/s41562-020-0898-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41562-020-0898-6
This article is cited by
-
Trends in the prevalence of social isolation among middle and older adults in China from 2011 to 2018: the China Health and Retirement Longitudinal Study
BMC Public Health (2024)
-
Knowledge diffusion in social networks under targeted attack and random failure: the resilience of communities
Journal of Economic Interaction and Coordination (2024)
-
Reproduction number projection for the COVID-19 pandemic
Advances in Continuous and Discrete Models (2023)
-
Quantifying the spatial spillover effects of non-pharmaceutical interventions on pandemic risk
International Journal of Health Geographics (2023)
-
Modelling the reopen strategy from dynamic zero-COVID in China considering the sequela and reinfection
Scientific Reports (2023)