Abstract
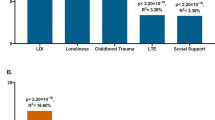
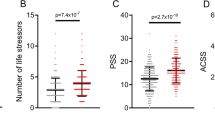
Advancing ability to predict who is likely to develop depression holds great potential in reducing the disease burden. Here, we use the predictable and large increase in depression with physician training stress to identify predictors of depression. Applying the major depressive disorder polygenic risk score (MDD-PRS) derived from the most recent Psychiatric Genomics Consortium–UK Biobank–23andMe genome-wide association study to 5,227 training physicians, we found that MDD-PRS predicted depression under training stress (β = 0.095, P = 4.7 × 10−16) and that MDD-PRS was more strongly associated with depression under stress than at baseline (MDD-PRS × stress interaction β = 0.036, P = 0.005). Further, known risk factors accounted for substantially less of the association between MDD-PRS and depression when under stress than at baseline, suggesting that MDD-PRS adds unique predictive power in depression prediction. Finally, we found that low MDD-PRS may have particular use in identifying individuals with high resilience. Together, these findings suggest that MDD-PRS holds promise in furthering our ability to predict vulnerability and resilience under stress.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The de-identified data from Intern Health Study are available through the PGC: https://www.med.unc.edu/pgc/shared-methods.
PGC phase 2–UK Biobank–23andMe MDD GWAS meta-analysis summary statistics: https://www.nature.com/articles/s41593-018-0326-7#data-availability.
Code availability
Custom code that supports the findings of this study is available from the corresponding author upon request.
References
Friedrich, M. J. Depression is the leading cause of disability around the world. J. Am. Med. Assoc. 317, 1517 (2017).
Rush, A. J. et al. Bupropion-SR, sertraline or venlafaxine-XR after failure of SSRIs for depression. N. Engl. J. Med. 354, 1231–1242 (2006).
Rossen, L. M., Hedegaard, H., Khan, D. & Warner, M. County-level trends in suicide rates in the U.S., 2005–2015. Am. J. Prev. Med. 55, 72–79 (2018).
Committee on Prevention of Mental Disorders & Institute of Medicine. Reducing Risks for Mental Disorders: Frontiers for Preventive Intervention Research (National Academies Press, 1994).
O’Connell, M. E. et al. (eds) Preventing Mental, Emotional, and Behavioral Disorders Among Young People: Progress and Possibilities (National Academies Press, 2009).
Prendes-Alvarez, S. & Nemeroff, C. B. Personalized medicine: prediction of disease vulnerability in mood disorders. Neurosci. Lett. 669, 10–13 (2018).
Sullivan, P. F., Neale, M. C. & Kendler, K. S. Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry 157, 1552–1562 (2000).
Okbay, A. et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016).
CONVERGE Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591 (2015).
Hyde, C. L. et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 48, 1031–1036 (2016).
Howard, D. M. et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 9, 1470 (2018).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013).
Chatterjee, N., Shi, J. & García-Closas, M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 17, 392–406 (2016).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
Kessler, R. C. The effects of stressful life events on depression. Annu. Rev. Psychol. 48, 191–214 (1997).
Maciejewski, P. K. & Mazure, C. M. Stressful life events and depression. Am. J. Psychiatry 157, 1344–1345 (2000).
Mata, D. A. et al. Prevalence of depression and depressive symptoms among resident physicians: a systematic review and meta-analysis. J. Am. Med. Assoc. 314, 2373–2383 (2015).
Sen, S. et al. A prospective cohort study investigating factors associated with depression during medical internship. Arch. Gen. Psychiatry 67, 557–565 (2010).
Martin, A. R. et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 100, 635–649 (2017).
Navrady, L. B. et al. Genetic risk of major depressive disorder: the moderating and mediating effects of neuroticism and psychological resilience on clinical and self-reported depression. Psychol. Med. 48, 1890–1899 (2018).
Middeldorp, C. M. & Wray, N. R. The value of polygenic analyses in psychiatry. World Psychiatry 17, 26–28 (2018).
Guille, C. et al. Web-based cognitive behavioral therapy intervention for the prevention of suicidal ideation in medical interns: a randomized clinical trial. JAMA Psychiatry 72, 1192–1198 (2015).
Southwick, S. M. & Charney, D. S. The science of resilience: implications for the prevention and treatment of depression. Science 338, 79–82 (2012).
Harper, A. R., Nayee, S. & Topol, E. J. Protective alleles and modifier variants in human health and disease. Nat. Rev. Genet. 16, 689–701 (2015).
Diagnostic and Statistical Manual of Mental Disorders 5th edn (American Psychiatric Association, 2003).
Kroenke, K., Spitzer, R. L. & Williams, J. B. The PHQ-9: validity of a brief depression severity measure. J. Gen. Intern. Med. 16, 606–613 (2001).
Spitzer, R. L., Kroenke, K. & Williams, J. B. W. Validation and utility of a self-report version of PRIME-MD: the PHQ primary care study. J. Am. Med. Soc. 282, 1737–1744 (1999).
Levis, B., Benedetti, A. & Thombs, B. D. Accuracy of Patient Health Questionnaire-9 (PHQ-9) for screening to detect major depression: individual participant data meta-analysis. Br. Med. J. 365, l1476 (2019).
Costa, P. T. Jr & McCrae, R. R. Stability and change in personality assessment: the revised NEO Personality Inventory in the year 2000. J. Pers. Assess. 68, 86–94 (1997).
Taylor, S. E. et al. Early family environment, current adversity, the serotonin transporter promoter polymorphism, and depressive symptomatology. Biol. Psychiatry 60, 671–676 (2006).
Rogers, N. L., Cole, S. A., Lan, H.-C., Crossa, A. & Demerath, E. W. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am. J. Hum. Biol. 19, 319–326 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015).
Ware, E. B. et al. Heterogeneity in polygenic scores for common human traits. Preprint at bioRxiv https://doi.org/10.1101/106062 (2017).
Choi, K. W. et al. Prospective study of polygenic risk, protective factors, and incident depression following combat deployment in US Army soldiers. Psychol. Med. https://doi.org/10.1017/S0033291719000527 (2019).
Peyrot, W. J. et al. Effect of polygenic risk scores on depression in childhood trauma. Br. J. Psychiatry 205, 113–119 (2014).
Musliner, K. L. et al. Polygenic risk, stressful life events and depressive symptoms in older adults: a polygenic score analysis. Psychol. Med. 45, 1709–1720 (2015).
Tingley, D., Yamamoto, T., Hirose, K., Keele, L. & Imai, K. mediation: R package for causal mediation analysis. J. Stat. Softw. 59, 1–38 (2014).
Acknowledgements
We thank the training physicians for taking part in this study. We also thank the participants and researchers of PGC and UK Biobank, and the research participants and employees of 23andMe, for making this work possible. This project was funded by the National Institute of Mental Health (grant no. R01MH101459 to S.S.). The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.S. designed the study. S.S. and Y.F. developed the research question. Y.F. performed the data management and analysis. Y.F. and S.S. wrote the manuscript. L.S., P.S. and M.B. provided critical review, discussion and revision of the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Mary Elizabeth Sutherland.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1
Associations of Imputed Data Derived MDD Polygenic Risk Score with PHQ-9 Depressive Symptom Scores (N = 5,227).
Extended Data Fig. 2 Baseline and Internship PHQ-9 Depressive Symptom Scores by MDD-PRS Group.
5,227 Subjects from Intern Health Study were binned into 40 groups of 2.5% of subjects (n=131 per group) from low to high MDD-PRS (left to right). The 40 x-axis groups are defined by group-wise average standardized MDD-PRS. Average PHQ-9 score of each group at baseline (cyan dots) and during internship (orange dots) are plotted with 95% CI error bars. LOESS fitting line (dash line) shadowed by 95% CI and linear regression fitting line (solid line) were applied to both baseline and internship plots. Optimal span parameter for LOESS regression was selected by generalized cross-validation method.
Extended Data Fig. 3 Population Structure Based on the Top Two Principal Component (PC) Analysis of the Intern Health Study.
Blue, red and green boxes depicted the analysis inclusion range of European, South Asian and East Asian Groups.
Supplementary information
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Rights and permissions
About this article
Cite this article
Fang, Y., Scott, L., Song, P. et al. Genomic prediction of depression risk and resilience under stress. Nat Hum Behav 4, 111–118 (2020). https://doi.org/10.1038/s41562-019-0759-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41562-019-0759-3
This article is cited by
-
Genetic susceptibility and lifestyle modify the association of long-term air pollution exposure on major depressive disorder: a prospective study in UK Biobank
BMC Medicine (2023)
-
Prevalence and risk factors for depression among training physicians in China and the United States
Scientific Reports (2022)
-
Assessing the joint effects of brain aging and gut microbiota on the risks of psychiatric disorders
Brain Imaging and Behavior (2022)
-
A Gene-Environment Interaction Study of Polygenic Scores and Maltreatment on Childhood ADHD
Research on Child and Adolescent Psychopathology (2022)
-
Genomic heterogeneity affects the response to Daylight Saving Time
Scientific Reports (2021)