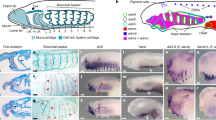
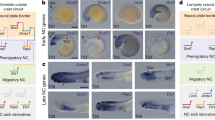
Abstract
Neural-crest cells and neuromesodermal progenitors (NMPs) are multipotent cells that are important for development of vertebrate embryos. In embryos of ascidians, which are the closest invertebrate relatives of vertebrates, several cells located at the border between the neural plate and the epidermal region have neural-crest-like properties; hence, the last common ancestor of ascidians and vertebrates may have had ancestral cells similar to neural-crest cells. However, these ascidian neural-crest-like cells do not produce cells that are commonly of mesodermal origin. Here we showed that a cell population located in the lateral region of the neural plate has properties resembling those of vertebrate neural-crest cells and NMPs. Among them, cells with Tbx6-related expression contribute to muscle near the tip of the tail region and cells with Sox1/2/3 expression give rise to the nerve cord. These observations and cross-species transcriptome comparisons indicate that these cells have properties similar to those of NMPs. Meanwhile, transcription factor genes Dlx.b, Zic-r.b and Snai, which are reminiscent of a gene circuit in vertebrate neural-crest cells, are involved in activation of Tbx6-related.b. Thus, the last common ancestor of ascidians and vertebrates may have had cells with properties of neural-crest cells and NMPs and such ancestral cells may have produced cells commonly of ectodermal and mesodermal origins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data generated during this study are included in this article and Zenodo (https://doi.org/10.5281/zenodo.10682771)81.
Code availability
No custom-made programmes were used in the present study. R-markdowns for analyses using Seurat are available from Zenodo (https://doi.org/10.5281/zenodo.10682771)81.
References
Tang, W. Y. & Bronner, M. E. Neural crest lineage analysis: from past to future trajectory. Development 147, dev193193 (2020).
Zalc, A. et al. Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 371, eabb4776 (2021).
Buitrago-Delgado, E., Nordin, K., Rao, A., Geary, L. & LaBonne, C. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science 348, 1332–1335 (2015).
Gans, C. & Northcutt, R. G. Neural crest and the origin of vertebrates—a new head. Science 220, 268–273 (1983).
Simoes-Costa, M. & Bronner, M. E. Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257 (2015).
Abitua, P. B., Wagner, E., Navarrete, I. A. & Levine, M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 492, 104–107 (2012).
Stolfi, A., Ryan, K., Meinertzhagen, I. A. & Christiaen, L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 527, 371–374 (2015).
Waki, K., Imai, K. S. & Satou, Y. Genetic pathways for differentiation of the peripheral nervous system in ascidians. Nat. Commun. 6, 8719 (2015).
Horie, R. et al. Shared evolutionary origin of vertebrate neural crest and cranial placodes. Nature 560, 228–232 (2018).
Papadogiannis, V. et al. Hmx gene conservation identifies the origin of vertebrate cranial ganglia. Nature 605, 701–705 (2022).
Tzouanacou, E., Wegener, A., Wymeersch, F. J., Wilson, V. & Nicolas, J. F. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell 17, 365–376 (2009).
Martin, B. L. & Kimelman, D. Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev. Cell 22, 223–232 (2012).
Henrique, D., Abranches, E., Verrier, L. & Storey, K. G. Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875 (2015).
Olivera-Martinez, I., Harada, H., Halley, P. A. & Storey, K. G. Loss of FGF-dependent mesoderm identity and rise of endogenous retinoid signalling determine cessation of body axis elongation. PLoS Biol. 10, e1001415 (2012).
Gouti, M., Metzis, V. & Briscoe, J. The route to spinal cord cell types: a tale of signals and switches. Trends Genet. 31, 282–289 (2015).
Takemoto, T. et al. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470, 394–398 (2011).
Goto, H., Kimmey, S. C., Row, R. H., Matus, D. Q. & Martin, B. L. FGF and canonical Wnt signaling cooperate to induce paraxial mesoderm from tailbud neuromesodermal progenitors through regulation of a two-step epithelial to mesenchymal transition. Development 144, 1412–1424 (2017).
Martin, B. L. & Steventon, B. A fishy tail: insights into the cell and molecular biology of neuromesodermal cells from zebrafish embryos. Dev. Biol. 487, 67–73 (2022).
Nishida, H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev. Biol. 121, 526–541 (1987).
Nicol, D. & Meinertzhagen, I. A. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. I. The early lineages of the neural plate. Dev. Biol. 130, 721–736 (1988).
Nicol, D. & Meinertzhagen, I. A. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. II. Neural plate morphogenesis and cell lineages during neurulation. Dev. Biol. 130, 737–766 (1988).
Mazet, F. et al. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev. Biol. 282, 494–508 (2005).
Wada, H., Saiga, H., Satoh, N. & Holland, P. W. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development 125, 1113–1122 (1998).
Abitua, P. B. et al. The pre-vertebrate origins of neurogenic placodes. Nature 524, 462–465 (2015).
Ikeda, T., Matsuoka, T. & Satou, Y. A time delay gene circuit is required for palp formation in the ascidian embryo. Development 140, 4703–4708 (2013).
Liu, B. & Satou, Y. Foxg specifies sensory neurons in the anterior neural plate border of the ascidian embryo. Nat. Commun. 10, 4911 (2019).
Wagner, E. & Levine, M. FGF signaling establishes the anterior border of the Ciona neural tube. Development 139, 2351–2359 (2012).
Hudson, C. & Yasuo, H. Neuromesodermal lineage contribution to CNS development in invertebrate and vertebrate chordates. Genes 12, 592 (2021).
Yagi, K., Takatori, N., Satou, Y. & Satoh, N. Ci-Tbx6b and Ci-Tbx6c are key mediators of the maternal effect gene Ci-macho1 in muscle cell differentiation in Ciona intestinalis embryos. Dev. Biol. 282, 535–549 (2005).
Imai, K. S., Hino, K., Yagi, K., Satoh, N. & Satou, Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047–4058 (2004).
Satou, Y., Kawashima, T., Shoguchi, E., Nakayama, A. & Satoh, N. An integrated database of the ascidian, Ciona intestinalis: towards functional genomics. Zool. Sci. 22, 837–843 (2005).
Satou, Y. et al. Gene expression profiles in Ciona intestinalis tailbud embryos. Development 128, 2893–2904 (2001).
Satou, Y., Imai, K. & Satoh, N. The ascidian Mesp gene specifies heart precursor cells. Development 131, 2533–2541 (2004).
Cao, C. et al. Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349–354 (2019).
Roure, A. & Darras, S. Msxb is a core component of the genetic circuitry specifying the dorsal and ventral neurogenic midlines in the ascidian embryo. Dev. Biol. 409, 277–287 (2016).
Imai, K. S., Stolfi, A., Levine, M. & Satou, Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136, 285–293 (2009).
Aniello, F. et al. Identification and developmental expression of Ci-msxb: a novel homologue of Drosophila msh gene in Ciona intestinalis. Mech. Dev. 88, 123–126 (1999).
Yagi, K. & Makabe, K. W. Isolation of an early neural maker gene abundantly expressed in the nervous system of the ascidian, Halocynthia roretzi. Dev. Genes Evol. 211, 49–53 (2001).
Nakamura, M. J., Terai, J., Okubo, R., Hotta, K. & Oka, K. Three-dimensional anatomy of the Ciona intestinalis tailbud embryo at single-cell resolution. Dev. Biol. 372, 274–284 (2012).
Imai, K. S., Hikawa, H., Kobayashi, K. & Satou, Y. Tfap2 and Sox1/2/3 cooperatively specify ectodermal fates in ascidian embryos. Development 144, 33–37 (2017).
Irvine, S. Q., Cangiano, M. C., Millette, B. J. & Gutter, E. S. Non-overlapping expression patterns of the clustered Dll-A/B genes in the ascidian Ciona intestinalis. J. Exp. Zool. B 308, 428–441 (2007).
Miya, T. & Nishida, H. Expression pattern and transcriptional control of SoxB1 in embryos of the ascidian Halocynthia roretzi. Zool. Sci. 20, 59–67 (2003).
Imai, K. S., Levine, M., Satoh, N. & Satou, Y. Regulatory blueprint for a chordate embryo. Science 312, 1183–1187 (2006).
Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015).
Nicol, D. & Meinertzhagen, I. A. Cell counts and maps in the larval central nervous system of the ascidian Ciona intestinalis (L.). J. Comp. Neurol. 309, 415–429 (1991).
Dufour, H. D. et al. Precraniate origin of cranial motoneurons. Proc. Natl Acad. Sci. USA 103, 8727–8732 (2006).
Takahashi, T. & Holland, P. W. Amphioxus and ascidian Dmbx homeobox genes give clues to the vertebrate origins of midbrain development. Development 131, 3285–3294 (2004).
Yasuo, H. & Satoh, N. Function of vertebrate T gene. Nature 364, 582–583 (1993).
Satoh, N., Tagawa, K. & Takahashi, H. How was the notochord born? Evol. Dev. 14, 56–75 (2012).
Schock, E. N., York, J. R. & LaBonne, C. The developmental and evolutionary origins of cellular pluripotency in the vertebrate neural crest. Semin. Cell Dev. Biol. 138, 36–44 (2023).
York, J. R. & McCauley, D. W. The origin and evolution of vertebrate neural crest cells. Open Biol. 10, 190285 (2020).
Martik, M. L. & Bronner, M. E. Riding the crest to get a head: neural crest evolution in vertebrates. Nat. Rev. Neurosci. 22, 616–626 (2021).
Rothstein, M. & Simoes-Costa, M. On the evolutionary origins and regionalization of the neural crest. Semin. Cell Dev. Biol. 138, 28–35 (2023).
Green, S. A., Simoes-Costa, M. & Bronner, M. E. Evolution of vertebrates as viewed from the crest. Nature 520, 474–482 (2015).
Leung, A. W. et al. WNT/beta-catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development 143, 398–410 (2016).
Frith, T. J. et al. Human axial progenitors generate trunk neural crest cells in vitro. eLife 7, e35786 (2018).
Gomez, G. A. et al. WNT/β-catenin modulates the axial identity of embryonic stem cell-derived human neural crest. Development 146, dev175604 (2019).
Martik, M. L. et al. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 574, 675–678 (2019).
Rothstein, M., Bhattacharya, D. & Simoes-Costa, M. The molecular basis of neural crest axial identity. Dev. Biol. 444, S170–S180 (2018).
Satou, Y. et al. A manually curated gene model set for an ascidian, Ciona robusta (Ciona intestinalis Type A). Zool. Sci. 39, 253–260 (2022).
Ikeda, T. & Satou, Y. Differential temporal control of Foxa.a and Zic-r.b specifies brain versus notochord fate in the ascidian embryo. Development 144, 38–43 (2017).
Tokuoka, M., Kobayashi, K. & Satou, Y. Distinct regulation of Snail in two muscle lineages of the ascidian embryo achieves temporal coordination of muscle development. Development 145, dev163915 (2018).
Corbo, J. C., Levine, M. & Zeller, R. W. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589–602 (1997).
Leggio, B. et al. MorphoNet: an interactive online morphological browser to explore complex multi-scale data. Nat. Commun. 10, 2812 (2019).
Nishida, H. Cell-division pattern during gastrulation of the ascidian, Halocynthia roretzi. Dev. Growth Differ. 28, 191–201 (1986).
Roure, A., Lemaire, P. & Darras, S. An otx/nodal regulatory signature for posterior neural development in ascidians. PLoS Genet. 10, e1004548 (2014).
Satou, Y. et al. A nearly complete genome of Ciona intestinalis type A (C. robusta) reveals the contribution of inversion to chromosomal evolution in the genus Ciona. Genome Biol. Evol. 11, 3144–3157 (2019).
Tokuoka, M., Imai, K. S., Satou, Y. & Satoh, N. Three distinct lineages of mesenchymal cells in Ciona intestinalis embryos demonstrated by specific gene expression. Dev. Biol. 274, 211–224 (2004).
Kusakabe, T. et al. Gene expression profiles in tadpole larvae of Ciona intestinalis. Dev. Biol. 242, 188–203 (2002).
Hotta, K. et al. Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev. Biol. 224, 69–80 (2000).
Chiba, S. et al. A genomewide survey of developmentally relevant genes in Ciona intestinalis. IX. Genes for muscle structural proteins. Dev. Genes Evol. 213, 291–302 (2003).
Chiba, S., Satou, Y., Nishikata, T. & Satoh, N. Isolation and characterization of cDNA clones for epidermis-specific and muscle-specific genes in Ciona savignyi embryos. Zool. Sci. 15, 239–246 (1998).
Kusakabe, T. G. et al. A conserved non-reproductive GnRH system in chordates. PLoS ONE 7, e41955 (2012).
Kobayashi, K., Maeda, K., Tokuoka, M., Mochizuki, A. & Satou, Y. Controlling cell fate specification system by key genes determined from network structure. iScience 4, 281–293 (2018).
Katsuyama, Y. et al. Regulation of synaptotagmin gene expression during ascidian embryogenesis. Dev. Biol. 244, 293–304 (2002).
Imai, K., Takada, N., Satoh, N. & Satou, Y. β-catenin mediates the specification of endoderm cells in ascidian embryos. Development 127, 3009–3020 (2000).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Martin, F. J. et al. Ensembl 2023. Nucleic Acids Res. 51, D933–D941 (2023).
Ishida, T. & Satou, Y. Ascidian embryonic cells with properties of neural-crest cells and neuromesodermal progenitors of vertebrates. Zenodo https://zenodo.org/records/10682771 (2024).
Acknowledgements
We thank R. Masuda, S. Tokuhiro, C. Imaizumi (Kyoto University), M. Yoshida (University of Tokyo) and other members working under the National BioResource Project for Ciona (MEXT, Japan) at Kyoto University and the University of Tokyo for providing experimental animals. This research was supported by grants from the Japan Society for the Promotion of Science under the grant nos. 21H02486 and 21H05239 to Y.S. The manuscript was edited by a technical editor, S. D. Aird.
Author information
Authors and Affiliations
Contributions
T.I. and Y.S. conceived and designed the experiments. T.I. performed experiments. T.I. and Y.S. analysed the data. Y.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Joshua York and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Acta.a, which encodes a muscle actin, is expressed in b9.34 at the late gastrula stage.
Acta.a expression was examined by in situ hybridization (green). Tbx6-r.b expression was also examined (magenta). Z-projected image stacks overlaid in pseudocolor are shown in the top row. The sixth optical slice more clearly shows that Acta.a and Tbx6-r.b are expressed in b9.34, but not in the other LNPCs. The eleventh optical slice shows expression of Acta.a and Tbx6-r.b in A9.31. Brightness and contrast of photographs were linearly adjusted. Scale bars, 50 μm.
Extended Data Fig. 2 Some pLNPC-derived cells change location from the dorsal to the ventral side between early and late tailbud stages.
A tailbud embryo expressing Hebp-r.a>Kaede reporter was UV-irradiated for photoconversion of Kaede fluorescence at the early tailbud stage (0 min; early tailbud I). One cell with photoconverted Kaede changed its location to the ventral side. Note that only two tail-tip cells are labelled because of mosaic incorporation of the reporter construct. The first, third and sixth photographs are the same as the photograph shown in Fig. 2d. Photographs are z-projected image stacks overlaid in pseudocolor. Brightness and contrast of photographs were linearly adjusted. The dorsal side is up and the ventral side is down. Scale bar, 50 μm.
Extended Data Fig. 3 Expression of a pan-neural marker, Celf3.a, and an endodermal-strand marker, Slc39a-related, in tailbud embryos.
a,b, Expression of Celf3.a (a) and Slc39a-related (b) was examined by in situ hybridization and photographs are z-projected image stacks overlaid in pseudocolor (Celf3.a and Slc39a-related, green; Hebp-r.a, magenta). Tail-tip cells, which express Hebp-r.a, do not express Celf3.a or Slc39a-related. Brightness and contrast of photographs were linearly adjusted. Tail-tip cells are indicated by white arrows. Nerve-cord cells are indicated by cyan arrows and endodermal-strand cells are indicated by yellow arrows. There are two putative germ-line cells between anterior endodermal-strand cells and tail-tip cells (orange arrows). Grey arrows indicate epidermal cells. Nuclei are stained with DAPI (grey). Scale bar, 50 μm.
Extended Data Fig. 4 Dlx.b, Msx, Snai, Zic-r.b, Tfap2-r.b, Pax3/7, Ets1/2.b, Lmx1 and Id.b are expressed in LNPCs.
(a–i) Expression was examined by in situ hybridization and photographs are z-projected image stacks and overlaid in pseudocolor (magenta). Nuclei were stained with DAPI (grey). Higher magnification views are shown on the right. Arrowheads indicate gene expression and arrows indicate the absence of expression. Developmental stages are shown in photographs; eG, early gastrula; mG, middle gastrula; eN, early neurula. Scale bar, 50 μm. (j) A summary of gene expression in LNPCs at the early and middle gastrula stages.
Extended Data Fig. 5 Tbx6-r.a expression in a middle gastrula embryo.
Tbx6-r.a is expressed in pLNPCs (b9.33 and b9.34) and indicated by white arrowheads. The remaining LNPCs (aLNPCs) are shown by arrows. Photographs are z-projected image stacks overlaid in pseudocolor. Nuclei are stained with DAPI (grey) and a higher magnification view is shown on the right. Brightness and contrast of photographs were linearly adjusted. Scale bars, 50 μm.
Extended Data Fig. 6 Expression of Sox1/2/3 in unperturbed middle gastrula embryos.
(a, b) Sox1/2/3 mRNA is detected in cells including LNPCs (white arrowheads) of normal middle gastrula gastrula (mG) and early neurula (eN) embryos. Note that signals in pLNPCs (b9.33 and b9.34) are weak at the early neurula stage. (c) Sox1/2/3 nascent transcripts, which were examined with an intron probe, are not seen in pLNPCs (white arrows), which indicates that Sox1/2/3 is not transcribed in pLNPCs. Note that Sox1/2/3 is expressed in cells that contribute to the central nervous system (grey arrowheads) and aLNPCs (white arrowheads). Photographs are z-projected image stacks overlaid in pseudocolor; magenta, in situ hybridization signals; grey, nuclei stained with DAPI. Brightness and contrast of photographs were linearly adjusted. Scale bar, 50 μm.
Extended Data Fig. 7 Expression of Sox1/2/3 in morphant embryos of Msx, Snai, Dlx.b, or Zic-r.b at the middle gastrula stage.
Expression of Sox1/2/3 nascent transcripts was examined by in situ hybridization using a probe designed to hybridize with the first intron of Sox1/2/3. Higher magnification views are shown on the right. LNPCs that express or do not express designated genes are shown by arrowheads and arrows, respectively. Nuclei are stained with DAPI and are shown in grey. Photographs are z-projected image stacks overlaid in pseudocolor. Brightness and contrast of photographs were linearly adjusted. Numbers of embryos examined and embryos that expressed Sox1/2/3 nascent transcripts in b9.37 and b9.38 are shown in the panels. Scale bars, 50 μm.
Extended Data Fig. 8 High-resolution clustering of single-cell transcriptome data of Ciona and zebrafish.
High-resolution clustering results are mapped on the same UMAP plot that is shown in Fig. 5. Different clusters are indicated by different colours. Pie charts show that notochord and muscle cells of Ciona and zebrafish are largely grouped into the same clusters.
Extended Data Fig. 9 Low-resolution clustering of single-cell transcriptome data of Ciona and zebrafish.
Low-resolution clustering results are mapped on the same UMAP plot that is shown in Fig. 5. Different clusters are indicated by different colours. The pie chart shows that zebrafish cells annotated ‘tailbud presomitic mesoderm’ are mostly in clusters 6 and 10.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4.
Supplementary Tables
Supplementary Tables 1–11.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ishida, T., Satou, Y. Ascidian embryonic cells with properties of neural-crest cells and neuromesodermal progenitors of vertebrates. Nat Ecol Evol (2024). https://doi.org/10.1038/s41559-024-02387-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-024-02387-8
This article is cited by
-
Ascidian embryonic cells with properties of neural-crest cells and neuromesodermal progenitors of vertebrates
Nature Ecology & Evolution (2024)