Abstract
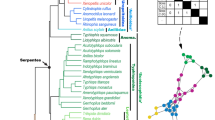
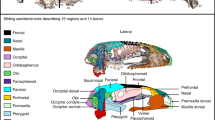
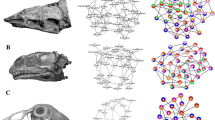
The arrangement and morphology of the vertebrate skull reflect functional and ecological demands, making it a highly adaptable structure. However, the fundamental developmental and macroevolutionary mechanisms leading to different vertebrate skull phenotypes remain unclear. Here we exploit the morphological diversity of squamate reptiles to assess the developmental and evolutionary patterns of skull variation and covariation in the whole head. Our geometric morphometric analysis of a complex squamate ontogenetic dataset (209 specimens, 169 embryos, 44 species), covering stages from craniofacial primordia to fully ossified bones, reveals that morphological differences between snake and lizard skulls arose gradually through changes in spatial relationships (heterotopy) followed by alterations in developmental timing or rate (heterochrony). Along with dynamic spatiotemporal changes in the integration pattern of skull bone shape and topology with surrounding brain tissues and sensory organs, we identify a relatively higher phenotypic integration of the developing snake head compared with lizards. The eye, nasal cavity and Jacobson’s organ are pivotal in skull morphogenesis, highlighting the importance of sensory rearrangements in snake evolution. Furthermore, our findings demonstrate the importance of early embryonic, ontogenetic and tissue interactions in shaping craniofacial evolution and ecological diversification in squamates, with implications for the nature of cranio-cerebral relations across vertebrates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw landmark data, encompassing all the required landmarks needed for replicating the analyses, and surface files used for visualization are available on the Zenodo repository (https://zenodo.org/record/8376575). CT-scan data for key adult squamate species and the embryonic series of L. lugubris are publicly available in the published literature53,88 and Morphosource database (https://www.morphosource.org/), respectively.
Code availability
R scripts for conducting the analyses and surface files used in specific analyses are available on the Zenodo repository (https://zenodo.org/record/8376575).
References
Hirasawa, T. & Kuratani, S. Evolution of the vertebrate skeleton: morphology, embryology, and development. Zool. Lett. 1, 2 (2015).
Emerson, S. B. & Bramble, D. M. in The Skull: Functional and Evolutionary Mechanisms Vol. 3 (eds Hanken, J. & Hall, B. K.) 384–421 (University of Chicago Press, 1993).
Patterson, M. et al. Ontogenetic shift in diet of a large elapid snake is facilitated by allometric change in skull morphology. Evol. Ecol. 36, 489–509 (2022).
Scanferla, A. Postnatal ontogeny and the evolution of macrostomy in snakes. R. Soc. Open Sci. 3, 160612 (2016).
Schwab, J. A. et al. Ontogenetic variation in the crocodylian vestibular system. J. Anat. 240, 821–832 (2022).
Esquerré, D., Sherratt, E. & Keogh, J. S. Evolution of extreme ontogenetic allometric diversity and heterochrony in pythons, a clade of giant and dwarf snakes. Evolution 71, 2829–2844 (2017).
Pavón-Vázquez, C. J., Esquerré, D. & Keogh, J. S. Ontogenetic drivers of morphological evolution in monitor lizards and allies (Squamata: Paleoanguimorpha), a clade with extreme body size disparity. BMC Ecol. Evol. 22, 15 (2022).
Fernandez Blanco, M. V., Cassini, G. H. & Bona, P. Skull ontogeny of extant caimans: a three-dimensional geometric morphometric approach. Zoology 129, 69–81 (2018).
Foth, C., Hedrick, B. P. & Ezcurra, M. D. Cranial ontogenetic variation in early saurischians and the role of heterochrony in the diversification of predatory dinosaurs. PeerJ 4, e1589 (2016).
Palci, A., Lee, M. S. Y. & Hutchinson, M. N. Patterns of postnatal ontogeny of the skull and lower jaw of snakes as revealed by micro-CT scan data and three-dimensional geometric morphometrics. J. Anat. 229, 723–754 (2016).
Piras, P. et al. The role of post-natal ontogeny in the evolution of phenotypic diversity in Podarcis lizards: Podarcis lizards post-natal ontogeny. J. Evol. Biol. 24, 2705–2720 (2011).
Geiger, M. et al. Neomorphosis and heterochrony of skull shape in dog domestication. Sci. Rep. 7, 13443 (2017).
Da Silva, F. O. et al. The ecological origins of snakes as revealed by skull evolution. Nat. Commun. 9, 376 (2018).
Fabbri, M. et al. The skull roof tracks the brain during the evolution and development of reptiles including birds. Nat. Ecol. Evol. 1, 1543–1550 (2017).
Bhullar, B.-A. S. et al. Birds have paedomorphic dinosaur skulls. Nature 487, 223–226 (2012).
Morris, Z. S., Vliet, K. A., Abzhanov, A. & Pierce, S. E. Heterochronic shifts and conserved embryonic shape underlie crocodylian craniofacial disparity and convergence. Proc. Biol. Sci. 286, 20182389 (2019).
Morris, Z. S., Vliet, K. A., Abzhanov, A. & Pierce, S. E. Developmental origins of the crocodylian skull table and platyrostral face. Anat. Rec. 305, 2838–2853 (2022).
Navalón, G. et al. Craniofacial development illuminates the evolution of nightbirds (Strisores). Proc. Biol. Sci. 288, 20210181 (2021).
Fabbri, M. et al. A shift in ontogenetic timing produced the unique sauropod skull. Evolution 75, 819–831 (2021).
Goswami, A., Polly, P. D., Mock, O. B. & Sánchez-Villagra, M. R. Shape, variance and integration during craniogenesis: contrasting marsupial and placental mammals. J. Evol. Biol. 25, 862–872 (2012).
Gray, J. A., Sherratt, E., Hutchinson, M. N. & Jones, M. E. H. Changes in ontogenetic patterns facilitate diversification in skull shape of Australian agamid lizards. BMC Evol. Biol. 19, 1–10 (2019).
Evans, K. M., Waltz, B., Tagliacollo, V., Chakrabarty, P. & Albert, J. S. Why the short face? Developmental disintegration of the neurocranium drives convergent evolution in neotropical electric fishes. Ecol. Evol. 7, 1783–1801 (2017).
Colangelo, P., Ventura, D., Piras, P., Pagani Guazzugli Bonaiuti, J. & Ardizzone, G. Are developmental shifts the main driver of phenotypic evolution in Diplodus spp. (Perciformes: Sparidae)? BMC Evol. Biol. 19, 106 (2019).
White, H. E. et al. Pedomorphosis in the ancestry of marsupial mammals. Curr. Biol. 33, 2136–2150.e4 (2023).
Wilson, L. A. B. et al. Patterns of ontogenetic evolution across extant marsupials reflect different allometric pathways to ecomorphological diversity. Nat. Commun. 14, 2689 (2023).
Young, N. M. et al. Embryonic bauplans and the developmental origins of facial diversity and constraint. Development 141, 1059–1063 (2014).
Griffin, C. T. et al. The developing bird pelvis passes through ancestral dinosaurian conditions. Nature 608, 346–352 (2022).
Beeching, S. C., Elsey, R. M. & Rehorek, S. J. Ontogeny of the American alligator (Alligator mississippiensis) prenatal head: a morphometric approach. J. Morphol. 283, 805–814 (2022).
Navalón, G., Marugán-Lobón, J., Bright, J. A., Cooney, C. R. & Rayfield, E. J. The consequences of craniofacial integration for the adaptive radiations of Darwin’s finches and Hawaiian honeycreepers. Nat. Ecol. Evol. 4, 270–278 (2020).
Hedrick, B. P. et al. Morphological diversification under high integration in a hyper diverse mammal clade. J. Mamm. Evol. 27, 563–575 (2020).
Goswami, A., Smaers, J. B., Soligo, C. & Polly, P. D. The macroevolutionary consequences of phenotypic integration: from development to deep time. Phil. Trans. R. Soc. B 369, 20130254 (2014).
Ackermann, R. Ontogenetic integration of the hominoid face. J. Hum. Evol. 48, 175–197 (2005).
Hanson, M., Hoffman, E. A., Norell, M. A. & Bhullar, B.-A. S. The early origin of a birdlike inner ear and the evolution of dinosaurian movement and vocalization. Science 372, 601–609 (2021).
Yi, H. & Norell, M. A. The burrowing origin of modern snakes. Sci. Adv. 1, e1500743 (2015).
Evers, S. W. et al. Independent origin of large labyrinth size in turtles. Nat. Commun. 13, 5807 (2022).
Lautenschlager, S., Ferreira, G. S. & Werneburg, I. Sensory evolution and ecology of early turtles revealed by digital endocranial reconstructions. Front. Ecol. Evol. 6, 7 (2018).
DeMyer, W., Zeman, W. & Palmer, C. G. The face predicts the brain: diagnostic significance of median facial anomalies for holoprosencephaly (arhinencephaly). Pediatrics 34, 256–263 (1964).
Ferreira, G. S., Werneburg, I., Lautenschlager, S. & Evers, S. W. in Paleoneurology of Amniotes: New Directions in the Study of Fossil Endocasts (eds. Dozo, M. T., Paulina-Carabajal, A., Macrini, T. E. & Walsh, S.) 79–121 (Springer International Publishing, 2023).
Koyabu, D. et al. Mammalian skull heterochrony reveals modular evolution and a link between cranial development and brain size. Nat. Commun. 5, 3625 (2014).
Marugán‐Lobón, J., Nebreda, S. M., Navalón, G. & Benson, R. B. J. Beyond the beak: brain size and allometry in avian craniofacial evolution. J. Anat. 240, 197–209 (2022).
Richtsmeier, J. T. et al. Phenotypic integration of neurocranium and brain. J. Exp. Zool. B 306, 360–378 (2006).
Hu, D. et al. Signals from the brain induce variation in avian facial shape. Dev. Dyn. 244, 1133–1143 (2015).
Martínez-Abadías, N. et al. FGF/FGFR signaling coordinates skull development by modulating magnitude of morphological integration: evidence from Apert syndrome mouse models. PLoS ONE 6, e26425 (2011).
Conith, A. J., Hope, S. A. & Albertson, R. C. Covariation of brain and skull shapes as a model to understand the roles for crosstalk in development and evolution. Evol. Dev. 25, 85–102 (2023).
Evans, S. E. in Biology of the Reptilia, Morphology H: The Skull of Lepidosauria Vol. 20 (eds. Gans, C., Gaunt, A. S. & Adler, K.) 2–227 (Society for the Study of Amphibians and Reptiles, 2008).
Cundall, D. & Irish, F. in Biology of the Reptilia, Morphology H: The Skull of Lepidosauria Vol. 20 (eds. Gans, C., Gaunt, A. S. & Adler, K.) 349–692 (Society for the Study of Amphibians and Reptiles, 2008).
Ollonen, J., Da Silva, F. O., Mahlow, K. & Di-Poï, N. Skull development, ossification pattern, and adult shape in the emerging lizard model organism Pogona vitticeps: a comparative analysis with other squamates. Front. Physiol. 9, 278 (2018).
Watanabe, A. et al. Ecomorphological diversification in squamates from conserved pattern of cranial integration. Proc. Natl Acad. Sci. USA 116, 14688–14697 (2019).
Werneburg, I. & Sánchez-Villagra, M. R. Skeletal heterochrony is associated with the anatomical specializations of snakes among squamate reptiles. Evolution 69, 254–263 (2015).
Rhoda, D., Polly, P. D., Raxworthy, C. & Segall, M. Morphological integration and modularity in the hyperkinetic feeding system of aquatic‐foraging snakes. Evolution 75, 56–72 (2021).
Asakura, Y. & Kawabe, S. Anatomical network analyses reveal evolutionary integration and modularity in the lizards skull. Sci. Rep. 12, 14429 (2022).
Strong, C. R. C., Scherz, M. D. & Caldwell, M. W. Convergence, divergence, and macroevolutionary constraint as revealed by anatomical network analysis of the squamate skull, with an emphasis on snakes. Sci. Rep. 12, 14469 (2022).
Macrì, S., Savriama, Y., Khan, I. & Di-Poï, N. Comparative analysis of squamate brains unveils multi-level variation in cerebellar architecture associated with locomotor specialization. Nat. Commun. 10, 5560 (2019).
Underwood, G. in Biology of the Reptilia, Morphology B Vol. 2 (eds. Gans, C. & Parsons, T. S.) 1–97 (Academic Press, 1970).
Schoch, R. R. Amphibian skull evolution: the developmental and functional context of simplification, bone loss and heterotopy. J. Exp. Zool. B 322, 619–630 (2014).
Tokita, M., Chaeychomsri, W. & Siruntawineti, J. Skeletal gene expression in the temporal region of the reptilian embryos: implications for the evolution of reptilian skull morphology. SpringerPlus 2, 336 (2013).
Lee, H. W., Esteve-Altava, B. & Abzhanov, A. Evolutionary and ontogenetic changes of the anatomical organization and modularity in the skull of archosaurs. Sci. Rep. 10, 16138 (2020).
Plateau, O. & Foth, C. Birds have peramorphic skulls, too: anatomical network analyses reveal oppositional heterochronies in avian skull evolution. Commun. Biol. 3, 195 (2020).
Fabre, A.-C. et al. Metamorphosis shapes cranial diversity and rate of evolution in salamanders. Nat. Ecol. Evol. 4, 1129–1140 (2020).
Evans, K. M. et al. Integration drives rapid phenotypic evolution in flatfishes. Proc. Natl Acad. Sci. USA 118, e2101330118 (2021).
Simões, T. R., Vernygora, O., Caldwell, M. W. & Pierce, S. E. Megaevolutionary dynamics and the timing of evolutionary innovation in reptiles. Nat. Commun. 11, 3322 (2020).
Sanger, T. J., Mahler, D. L., Abzhanov, A. & Losos, J. B. Roles for modularity and constraint in the evolution of cranial diversity among Anolis lizards. Evol. Int. J. Org. Evol. 66, 1525–1542 (2012).
Schwenk, K. Of tongues and noses: chemoreception in lizards and snakes. Trends Ecol. Evol. 10, 7–12 (1995).
Vidal, N. & Hedges, S. B. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C. R. Biol. 328, 1000–1008 (2005).
Segall, M., Cornette, R., Rasmussen, A. R. & Raxworthy, C. J. Inside the head of snakes: influence of size, phylogeny, and sensory ecology on endocranium morphology. Brain Struct. Funct. 226, 2401–2415 (2021).
Yaryhin, O., Klembara, J., Pichugin, Y., Kaucka, M. & Werneburg, I. Limb reduction in squamate reptiles correlates with the reduction of the chondrocranium: a case study on serpentiform anguids. Dev. Dyn. 250, 1300–1317 (2021).
Hsiang, A. Y. et al. The origin of snakes: revealing the ecology, behavior, and evolutionary history of early snakes using genomics, phenomics, and the fossil record. BMC Evol. Biol. 15, 87 (2015).
Gower, D. J., Hauzman, E., Simoes, B. F. & Schott, R. K. in The Origin and Early Evolutionary History of Snakes (eds. Gower, D. J. & Zaher, H.) 316–348 (Cambridge Univ. Press, 2022).
Yin, W. et al. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat. Commun. 7, 13107 (2016).
Castoe, T. A. et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl Acad. Sci. USA 110, 20645–20650 (2013).
Müller, J., Bickelmann, C. & Sobral, G. The evolution and fossil history of sensory perception in amniote vertebrates. Annu. Rev. Earth Planet. Sci. 46, 495–519 (2018).
Le Duc, D. & Schöneberg, T. Adaptation to nocturnality—learning from avian genomes. BioEssays 38, 694–703 (2016).
Caprette, C. L., Lee, M. S. Y., Shine, R., Mokany, A. & Downhower, J. F. The origin of snakes (Serpentes) as seen through eye anatomy. Biol. J. Linn. Soc. 81, 469–482 (2004).
Kishida, T. et al. Loss of olfaction in sea snakes provides new perspectives on the aquatic adaptation of amniotes. Proc. Biol. Sci. 286, 20191828 (2019).
Kishida, T. Olfaction of aquatic amniotes. Cell Tissue Res. 383, 353–365 (2021).
Brykczynska, U., Tzika, A. C., Rodriguez, I. & Milinkovitch, M. C. Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles. Genome Biol. Evol. 5, 389–401 (2013).
Gans, C. Tetrapod limblessness: evolution and functional corollaries. Am. Zool. 15, 455–467 (1975).
Wake, M. H. The comparative morphology and evolution of the eyes of caecilians (Amphibia, Gymnophiona). Zoomorphology 105, 277–295 (1985).
Cundall, D. & Rossman, D. A. Cephalic anatomy of the rare Indonesian snake Anomochilus weberi. Zool. J. Linn. Soc. 109, 235–273 (1993).
Strong, C. R. C., Palci, A. & Caldwell, M. W. Insights into skull evolution in fossorial snakes, as revealed by the cranial morphology of Atractaspis irregularis (Serpentes: Colubroidea). J. Anat. 238, 146–172 (2021).
Deckelbaum, R. A. et al. Regulation of cranial morphogenesis and cell fate at the neural crest-mesoderm boundary by engrailed 1. Development 139, 1346–1358 (2012).
Teng, C. S., Cavin, L., Maxson, R. E. Jr, Sánchez-Villagra, M. R. & Crump, J. G. Resolving homology in the face of shifting germ layer origins: lessons from a major skull vault boundary. eLife 8, e52814 (2019).
Maddin, H. C., Piekarski, N., Sefton, E. M. & Hanken, J. Homology of the cranial vault in birds: new insights based on embryonic fate-mapping and character analysis. R. Soc. Open Sci. 3, 160356 (2016).
Kuroda, S., Adachi, N. & Kuratani, S. A detailed redescription of the mesoderm/neural crest cell boundary in the murine orbitotemporal region integrates the mammalian cranium into a pan-amniote cranial configuration. Evol. Dev. 25, 32–53 (2023).
Noden, D. M. & Trainor, P. A. Relations and interactions between cranial mesoderm and neural crest populations. J. Anat. 207, 575–601 (2005).
Piekarski, N., Gross, J. B. & Hanken, J. Evolutionary innovation and conservation in the embryonic derivation of the vertebrate skull. Nat. Commun. 5, 5661 (2014).
Kague, E. et al. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE 7, e47394 (2012).
Griffing, A. H. et al. Embryonic development of a parthenogenetic vertebrate, the mourning gecko (Lepidodactylus lugubris). Dev. Dyn. 248, 1070–1090 (2019).
Werneburg, I., Polachowski, K. M. & Hutchinson, M. N. Bony skull development in the Argus monitor (Squamata, Varanidae, Varanus panoptes) with comments on developmental timing and adult anatomy. Zoology 118, 255–280 (2015).
Khannoon, E. R., Ollonen, J. & Di-Poï, N. Embryonic development of skull bones in the Sahara horned viper (Cerastes cerastes), with new insights into structures related to the basicranium and braincase roof. J. Anat. 237, 1–19 (2020).
Diaz, R. E. et al. Captive care, raising, and breeding of the veiled chameleon (Chamaeleo calyptratus). Cold Spring Harb. Protoc. 2015, 943–949 (2015).
Tahara, Y. & Obara, K. A novel shell-less culture system for chick embryos using a plastic film as culture vessels. J. Poult. Sci. 51, 307–312 (2014).
Klingenberg, C. P. MorphoJ: an integrated software package for geometric morphometrics. Mol. Ecol. Resour. 11, 353–357 (2010).
Baken, E. K., Collyer, M. L., Kaliontzopoulou, A. & Adams, D. C. geomorph v4.0 and gmShiny: enhanced analytics and a new graphical interface for a comprehensive morphometric experience. Methods Ecol. Evol. 12, 2355–2363 (2021).
Adams, D. C. & Otárola-Castillo, E. Geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4, 393–399 (2013).
Sievert, C. Interactive web-based data visualization with R, plotly, and shiny (Chapman and Hall/CRC, 2020).
Tonini, J. F. R., Beard, K. H., Ferreira, R. B., Jetz, W. & Pyron, R. A. Fully-sampled phylogenies of squamates reveal evolutionary patterns in threat status. Biol. Conserv. 204, 23–31 (2016).
Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinforma. 69, e96 (2020).
Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T.-Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017).
Yu, G., Lam, T. T.-Y., Zhu, H. & Guan, Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 35, 3041–3043 (2018).
Yu, G. Data Integration, Manipulation and Visualization of Phylogenetic Trees (Chapman and Hall/CRC, 2022).
Neuwirth, E. RColorBrewer: ColorBrewer Palettes. R version 1.1-3 https://cran.r-project.org/web/packages/RColorBrewer/index.html (2022).
Schlager, S. in Statistical Shape and Deformation Analysis (eds. Zheng, G., Li, S. & Székely, G.) 217–256 (Academic Press, 2017).
Antonio, P. et al. Arothron: an R package for geometric morphometric methods and virtual anthropology applications. Am. J. Phys. Anthropol. 176, 144–151 (2021).
Garnier, S. et al. sjmgarnier/viridis: viridis 0.6.0 (pre-CRAN release). Zenodo https://zenodo.org/records/7890878 (2021).
Collyer, M. L. & Adams, D. C. RRPP: an R package for fitting linear models to high‐dimensional data using residual randomization. Methods Ecol. Evol. 9, 1772–1779 (2018).
Collyer, M. & Adams, D. RRPP: linear model evaluation with randomized residuals in a permutation procedure. R version 1.3.1 https://github.com/mlcollyer/RRPP (2022).
Gerber, S. & Hopkins, M. J. Mosaic heterochrony and evolutionary modularity: the trilobite genus Zacanthopsis as a case study. Evolution 65, 3241–3252 (2011).
Marcy, A. E. et al. Australian rodents reveal conserved cranial evolutionary allometry across 10 million years of murid evolution. Am. Nat. 196, 755–768 (2020).
Werneburg, I. A standard system to study vertebrate embryos. PLoS ONE 4, e5887 (2009).
Lindeløv, J. K. mcp: an R package for regression with multiple change points. Preprint at OSF Preprints https://doi.org/10.31219/osf.io/fzqxv (2020).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Wickham, H., Vaughan, D. & Girlich, M. tidyr: tidy messy data. R version 1.3.0 https://tidyr.tidyverse.org/ (2023).
Wickham, H. et al. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019).
Wickham, H., François, R., Henry, L., Müller, K. & Vaughan, D. dplyr: a grammar of data manipulation. R version 1.1.3 https://dplyr.tidyverse.org/ (2023).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2021).
Watanabe, A., Balanoff, A. M., Gignac, P. M., Gold, M. E. L. & Norell, M. A. Novel neuroanatomical integration and scaling define avian brain shape evolution and development. eLife 10, e68809 (2021).
Adams, D. C. Evaluating modularity in morphometric data: challenges with the RV coefficient and a new test measure. Methods Ecol. Evol. 7, 565–572 (2016).
Epskamp, S., Cramer, A. O. J., Waldorp, L. J., Schmittmann, V. D. & Borsboom, D. qgraph: network visualizations of relationships in psychometric data. J. Stat. Softw. 48, 1–18 (2012).
Zeileis, A. & Hothorn, T. Diagnostic checking in regression relationships. R. News 2/3, 7–10 (2002).
Wood, S. N. Thin plate regression splines. J. R. Stat. Soc. Ser. B 65, 95–114 (2003).
Wood, S. N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models: estimation of semiparametric generalized linear models. J. R. Stat. Soc. Ser. B 73, 3–36 (2011).
Wood, S. N. Generalized Additive Models: An Introduction with R (Chapman and Hall/CRC, 2017).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Acknowledgements
We thank A.-C. Aho, M. Partanen, M. Snepere, T. Ahlskog and J. Ulpovaara for technical assistance in captive breeding and animal care; O. Ovaskainen and H. Laakkonen (Finnish Museum of Natural History) for specimen loans; Helsinki X-ray Laboratory, Department of Physics (University of Helsinki), and especially H. Suhonen and H. Help for access to X-ray CT facilities and assistance with scanning large samples; A. Griffing for providing access to Lepidodactylus lugubris CT data from www.MorphoSource.org (collection of the University of Florida, IP holder Marquette University, funded by National Science Foundation grants: Division of Environmental Biology, grant numbers 1657662 and 1657656, Division of Biological Infrastructure, grant number 1701714); J. Eymann and D. Razmadze for technical assistance; D. Esquerre for the code provided for the heterochrony analysis; K. Happonen for helpful assistance on the spline analysis; M. and P. Joki, Tropicario and LL Reptiles for the chameleon, python and corn snake eggs provided for this study; R. Johansson for a sample donation; K. Koponen, M. Launiainen, A. Seppälä, J. Jalkanen, T. Rissanen, M. Tiusanen, M. Aulio, P. Puustinen, M. P. Pulkkinen, J. Takkinen and L. Sagath for assistance in field work; all the landowners for the permits given for field work; members of the Di-Poï laboratory and R. Rice, L. Säilä-Corfe and J. Jernvall for helpful discussions; and D. Ho for proofreading. This work was supported by funds from the Academy of Finland (decision 321910 to N.D.-P.), Institute of Biotechnology (to N.D.-P.), Sigrid Jusélius Foundation (to N.D.-P.), Minerva Foundation (to N.D.-P.), Integrative Life Science Doctoral Program (to J.O.), Finnish Cultural Foundation (to J.O.), Kuopion Luonnon Ystävien Yhdistys (to J.O.), Societas Biologica Fennica Vanamo (to J.O.), Ella and Georg Ehrnrooth Foundation (to J.O.), Oskar Öflund Stiftelse sr (to J.O.), Doctoral School in Health Sciences (to J.O.), Swedish Cultural Foundation in Finland (to I.-M.A.), Carl Gans Foundation (to J.O.) and Deputyship for Research & lnnovation, Ministry of Education in Saudi Arabia (project number 445-5-702 to E.R.K.).
Author information
Authors and Affiliations
Contributions
J.O. and N.D.-P. designed and planned the overall experimental approach. J.O., E.R.K., S.M., V.V., J.K., J.S., A.S, I.W., R.E.D. and N.D.-P. contributed to embryonic and/or postnatal specimen collection and preparation. Micro-CT scans were carried out by J.O., S.M., J.K, A.S. and I.-M.A. The 3D reconstructions and segmentations were carried out by J.O. and S.M. J.O. collected the 3D landmark data and performed all geometric morphometric and statistical analyses. J.O. and N.D.-P. created the figures and wrote the article. All co-authors contributed in the form of discussion and critical comments. All authors approved the final version of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Reconstruction of ancestral trajectories.
Ancestral trajectories of modern snake (black) and toxicoferans (light grey) for skull (a, c, e) and soft tissues (b, d, f) at stages 1–2 (a, b), stage 3 (c, d), and stages 4–5 (e, f). The reconstructions involved the estimation of the ancestral intercept, angle, as well as minimum and maximum size. The five main developmental stages (stages 1–5) are colour-coded as before. Species names and the nodes employed for the reconstructions are indicated on the phylogenetic trees. Silhouettes from PhyloPic.org (creator credits: snake, Michael Keesay; lizard, Jose Carlos Arenas-Monroy).
Supplementary information
Supplementary Information
Supplementary Figs. 1–34, Tables 1–87 and References.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ollonen, J., Khannoon, E.R., Macrì, S. et al. Dynamic evolutionary interplay between ontogenetic skull patterning and whole-head integration. Nat Ecol Evol 8, 536–551 (2024). https://doi.org/10.1038/s41559-023-02295-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-023-02295-3