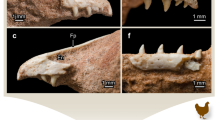
Abstract
Chondrichthyan dentitions are conventionally interpreted to reflect the ancestral gnathostome condition but interpretations of osteichthyan dental evolution in this light have proved unsuccessful, perhaps because chondrichthyan dentitions are equally specialized, or else evolved independently. Ischnacanthid acanthodians are stem-Chondrichthyes; as phylogenetic intermediates of osteichthyans and crown-chondrichthyans, the nature of their enigmatic dentition may inform homology and the ancestral gnathostome condition. Here we show that ischnacanthid marginal dentitions were statodont, composed of multicuspidate teeth added in distally diverging rows and through proximal superpositional replacement, while their symphyseal tooth whorls are comparable to chondrichthyan and osteichthyan counterparts. Ancestral state estimation indicates the presence of oral tubercles on the jaws of the gnathostome crown-ancestor; tooth whorls or tooth rows evolved independently in placoderms, osteichthyans, ischnacanthids, other acanthodians and crown-chondrichthyans. Crown-chondrichthyan dentitions are derived relative to the gnathostome crown-ancestor, which possessed a simple dentition and lacked a permanent dental lamina, which evolved independently in Chondrichthyes and Osteichthyes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data matrix is available at https://doi.org/10.6084/m9.figshare.14447139. Sources for taxa and age ranges and the phylogenetic character list are available as supplementary information. Tomograms and surface files are archived in the University of Bristol data repository, data.bris, at https://doi.org/10.5523/bris.1557rzkyzst5b2jagjuz9li5er.
Code availability
XML BEAST2 files, MrBayes Nexus files, BEAST1 XML files and R scripts are available at https://doi.org/10.6084/m9.figshare.14447139.
References
Smith, M. M. & Coates, M. I. Evolutionary origins of the vertebrate dentition: phylogenetic patterns and developmental evolution. Eur. J. Oral. Sci. 106, 482–500 (1998).
Botella, H., Blom, H., Dorka, M., Ahlberg, P. E. & Janvier, P. Jaws and teeth of the earliest bony fishes. Nature 448, 583–586 (2007).
Debiais-Thibaud, M. et al. Tooth and scale morphogenesis in shark: an alternative process to the mammalian enamel knot system. BMC Evol. Biol. 15, 292 (2015).
Rasch, L. J. et al. An ancient dental gene set governs development and continuous regeneration of teeth in sharks. Dev. Biol. 415, 347–370 (2016).
Smith, M. M., Fraser, G. J. & Mitsiadis, T. A. Dental lamina as source of odontogenic stem cells: evolutionary origins and developmental control of tooth generation in gnathostomes. J. Exp. Zool. B 312, 260–280 (2009).
Tucker, A. S. & Fraser, G. J. Evolution and developmental diversity of tooth regeneration. Semin. Cell Dev. Biol. 25, 71–80 (2014).
Coates, M. I. et al. An early chondrichthyan and the evolutionary assembly of a shark body plan. Proc. R. Soc. B 285, https://doi.org/10.1098/rspb.2017.2418 (2018).
Zhu, M. et al. A Silurian placoderm with osteichthyan-like marginal jaw bones. Nature 502, 188–193 (2013).
Giles, S., Friedman, M. & Brazeau, M. D. Osteichthyan-like cranial conditions in an Early Devonian stem gnathostome. Nature 520, 82–U175 (2015).
Smith, M. M. & Johanson, Z. Separate evolutionary origins of teeth from evidence in fossil jawed vertebrates. Science 299, 1235–1236 (2003).
Denison, R. H. Acanthodii (Gustav Fischer, 1979).
Smith, M. M. Vertebrate dentitions at the origin of jaws: when and how pattern evolved. Evol. Dev. 5, 394–413 (2003).
Blais, S. A., MacKenzie, L. A. & Wilson, M. V. H. Tooth-like scales in Early Devonian eugnathostomes and the ‘outside-in’ hypothesis for the origins of teeth in vertebrates. J. Vertebr. Paleontol. 31, 1189–1199 (2011).
Burrow, C. J., Newman, M., den Blaauwen, J., Jones, R. & Davidson, R. The Early Devonian ischnacanthiform acanthodian Ischnacanthus gracilis (Egerton, 1861) from the Midland Valley of Scotland. Acta Geol. Polon. 68, 335–362 (2018).
Burrow, C. J. Acanthodian fishes with dentigerous jaw bones: the Ischnacanthiformes and Acanthodopsis. Foss. Strat. 50, 8–22 (2004).
Lindley, I. D. Acanthodian fish remains from the lower devonian cavan bluff limestone (Murrumbidgee group), Taemas district, New South Wales. Alcheringa 24, 11–35 (2000).
Newman, M. J., Burrow, C. J. & den Blaaauwen, J. L. A new species of ischnacanthiform acanthodian from the Givetian of Mimerdalen, Svalbard. Norw. J. Geol. 99, 1–13 (2019).
Gross, W. Über das Gebiss der Acanthodier und Placodermen. Zool. J. Linn. Soc. 47, 121–130 (1967).
Ørvig, T. Acanthodian dentition and its bearing on the relationships of the group. Palaeontographica A 143, 119–150 (1973).
Smith, M. M. & Coates, M. I. in Major Events Of Early Vertebrate Evolution (ed. Ahlberg. P. E.) 223–240 (Taylor & Francis, 2001).
Donoghue, P. C. J. et al. Synchrotron X-ray tomographic microscopy of fossil embryos. Nature 442, 680–683 (2006).
Friedman, M. & Brazeau, M. D. A reappraisal of the origin and basal radiation of the osteichthyes. J. Vertebr. Paleontol. 30, 36–56 (2010).
Doeland, M., Couzens, A. M. C., Donoghue, P. C. J. & Rücklin, M. Tooth replacement in early sarcopterygians. R. Soc. Open Sci. 6, https://doi.org/10.1098/rsos.191173 (2019).
Jarvik, E. Middle and Upper Devonian porolepiformes from East Greenland with special reference to Glyptolepis groenlandica n. sp. and a discussion on the structure of the head of porolepiformes. Medd. Groenl. 187, 1–295 (1972).
Chen, D., Blom, H., Sanchez, S., Tafforeau, P. & Ahlberg, P. E. The stem osteichthyan Andreolepis and the origin of tooth replacement. Nature 539, 237–241 (2016).
Rücklin, M. et al. Development of teeth and jaws in the earliest jawed vertebrates. Nature 491, 748–751 (2012).
Clemen, G., Bartsch, P. & Wacker, K. Dentition and dentigerous bones in juveniles and adults of Polypterus senegalus (Cladistia, Actinopterygii). Ann. Anat. 180, 211–221 (1998).
Chen, D. et al. Development of cyclic shedding teeth from semi-shedding teeth: the inner dental arcade of the stem osteichthyan Lophosteus. R. Soc. Open Sci. 4, 161084 (2017).
Patterson, C. in Problems Of Phylogenetic Reconstruction (eds Joysey, K. A. & Friday, A. E.) Systematics Association Special Volume 21, 21–74 (Academic Press, 1982).
King, B., Qiao, T., Lee, M. S. Y., Zhu, M. & Long, J. A. Bayesian morphological clock methods resurrect placoderm monophyly and reveal rapid early evolution in jawed vertebrates. Syst. Biol. 66, 499–516 (2017).
Andreev, P. et al. The systematics of the Mongolepidida (Chondrichthyes) and the Ordovician origins of the clade. PeerJ 4, e1850 (2016).
Rücklin, M., Giles, S., Janvier, P. & Donoghue, P. C. J. Teeth before jaws? Comparative analysis of the structure and development of the external and internal scales in the extinct jawless vertebrate Loganellia scotica. Evol. Dev. 13, 523–532 (2011).
White, E. I. The Old Red Sandstone of Brown Lee Hill and the adjacent area. II. Palaeontology. Bull. Br. Mus. (Nat. Hist.) Geol. 5, 245–310 (1961).
Stampanoni, M. et al. TOMCAT: a beamline for tomographic microscopy and coherent radiology experiments. AIP Conf. Proc. 879, 848 (2007).
Maisey, J. G. et al. in Evolution and Development of FIshes (eds Johanson, Z., Underwood, C. J. & Richter, M.) 87–109 (Cambridge Univ. Press, 2018).
Bouckaert, R. et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15, e1006650 (2019).
Ayres, D. L. et al. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 61, 170–173 (2012).
Lewis, P. O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925 (2001).
Gavryushkina, A., Welch, D., Stadler, T. & Drummond, A. J. Bayesian inference of sampled ancestor trees for epidemiology and fossil calibration. PLoS Comput. Biol. 10, e1003919 (2014).
Drummond, A. J., Ho, S. Y. W., Phillips, M. J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, 699–710 (2006).
Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v1.6 http://beast.bio.ed.ac.uk/Tracer (2014).
Warren, D. L., Geneva, A. J. & Lanfear, R. RWTY (R We There Yet): an R package for examining convergence of Bayesian phylogenetic analyses. Mol. Biol. Evol. 34, 1016–1020 (2017).
King, B. Which morphological characters are influential in a Bayesian phylogenetic analysis? Examples from the earliest osteichthyans. Biol. Lett. 15, https://doi.org/10.1098/rsbl.2019.0288 (2019).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Bapst, D. W. paleotree: an R package for paleontological and phylogenetic analyses of evolution. Methods Ecol. Evol. 3, 803–807 (2012).
Brazeau, M. D. & Friedman, M. The characters of Palaeozoic jawed vertebrates. Zool. J. Linn. Soc. 170, 779–821 (2014).
Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, https://doi.org/10.1093/ve/vey016 (2018).
Lemey, P., Rambaut, A., Drummond, A. J. & Suchard, M. A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5, e1000520 (2009).
Minin, V. N. & Suchard, M. A. Counting labeled transitions in continuous-time Markov models of evolution. J. Math. Biol. 56, 391–412 (2008).
Xie, W., Lewis, P. O., Fan, Y., Kuo, L. & Chen, M. H. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst. Biol. 60, 150–160 (2011).
Kass, E. R. R. & Bayes, A. E. Factors. J. Am. Stat. Assoc. 90, 773–795 (1995).
Jombart, T. et al. OutbreakTools: a new platform for disease outbreak analysis using the R software. Epidemics 7, 28–34 (2014).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Schliep, K. P. phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011).
Acknowledgements
We thank S. Bengtson and D. Murdock for help at the TOMCAT beamline. We also thank E. Bernard (Natural History Museum) for access to collections and for facilitating the loan of specimens. The study was funded by an EU FP7 Marie-Curie Intra-European Fellowship (to M.R. and P.C.J.D.), Natural Environmental Research Council grant NE/G016623/1 (to P.C.J.D.) and Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) (VIDI 864.14.009 to M.R.). We acknowledge the Paul Scherrer Institut, Villigen, Switzerland, for provision of synchrotron radiation beamtime at the TOMCAT (X02DA) beamline of the Swiss Light Source (to P.C.J.D. and S. Bengtson).
Author information
Authors and Affiliations
Contributions
M.R. and P.C.J.D. designed the initial research. M.R., J.A.C., P.C.J.D. and F.M. performed scans. M.R. and J.A.C. segmented tomograms. B.K. produced the phylogenetic data matrix, and performed the phylogenetic analysis and ancestral state reconstruction. M.R. and P.C.J.D. drafted the manuscript, to which all authors contributed.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Ecology & Evolution thanks Gareth Fraser, Min Zhu, Moya Meredith Smith and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Virtual development of teeth on an ischnacanthid acanthodian jawbone.
Tooth rows of NRM-PZ P. 9449 Early Devonian, Canada. Labelled sclerochronology of the lateral row (a), lingual row (b) and overgrowth of the initial teeth at the centre of ossification (c). Colours of the nested boxes reflect the successive stages of tooth development. Scale bar represents 169 µm in a, b, and 72 µm in (c).
Extended Data Fig. 2 50% majority-rule consensus tree from tip-dated analysis of early gnathostome fossils.
‘Psarolepids’ constrained as stem osteichthyans, annotated with ancestral state reconstruction of tooth whorls.
Extended Data Fig. 3 Posterior probabilities from ancestral state reconstructions.
In column 1, ‘chondrichthyans’ refers to conventionally-defined chondrichthyans possessing tooth batteries. This includes Doliodus and crown chondrichthyans. Posterior probabilities are similar for tip-dated trees, and for undated Bayesian trees time-scaled a posteriori.
Supplementary information
Supplementary Information
Scores for taxa and ranges, character list and references.
Rights and permissions
About this article
Cite this article
Rücklin, M., King, B., Cunningham, J.A. et al. Acanthodian dental development and the origin of gnathostome dentitions. Nat Ecol Evol 5, 919–926 (2021). https://doi.org/10.1038/s41559-021-01458-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-021-01458-4
This article is cited by
-
Bony-fish-like scales in a Silurian maxillate placoderm
Nature Communications (2023)
-
Fossil evidence for a pharyngeal origin of the vertebrate pectoral girdle
Nature (2023)
-
The oldest gnathostome teeth
Nature (2022)