Abstract
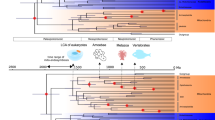
Though it is well accepted that mitochondria originated from an alphaproteobacteria-like ancestor, the phylogenetic relationship of the mitochondrial endosymbiont to extant Alphaproteobacteria is yet unresolved. The focus of much debate is whether the affinity between mitochondria and fast-evolving alphaproteobacterial lineages reflects true homology or artefacts. Approaches such as site exclusion have been claimed to mitigate compositional heterogeneity between taxa, but this comes at the cost of information loss, and the reliability of such methods is so far unproven. Here we demonstrate that site-exclusion methods produce erratic phylogenetic estimates of mitochondrial origin. Thus, previous phylogenetic hypotheses on the origin of mitochondria based on pretreated datasets should be re-evaluated. We applied alternative strategies to reduce phylogenetic noise by systematic taxon sampling while keeping site substitution information intact. Cross-validation based on a series of trees placed mitochondria robustly within Alphaproteobacteria, sharing an ancient common ancestor with Rickettsiales and currently unclassified marine lineages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The alignments and tree files generated in this study have been deposited in figshare (https://doi.org/10.6084/m9.figshare.12347216) (ref. 30).
Code availability
The script of the Bowker’s test score-based site-exclusion method is available as Supplementary Software.
References
Ku, C. et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524, 427–432 (2015).
Thiergart, T., Landan, G., Schenk, M., Dagan, T. & Martin, W. F. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol. Evol. 4, 466–485 (2012).
Abhishek, A., Bavishi, A., Bavishi, A. & Choudhary, M. Bacterial genome chimaerism and the origin of mitochondria. Can. J. Microbiol. 57, 49–61 (2011).
Atteia, A. et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26, 1533–1548 (2009).
Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Derelle, R. & Lang, B. F. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol. Biol. Evol. 29, 1277–1289 (2012).
Wang, Z. & Wu, M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 5, 7949 (2015).
Viklund, J., Ettema, T. J. & Andersson, S. G. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol. Biol. Evol. 29, 599–615 (2012).
Esser, C. et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 (2004).
Fitzpatrick, D. A., Creevey, C. J. & McInerney, J. O. Genome phylogenies indicate a meaningful alpha-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol. Biol. Evol. 23, 74–85 (2006).
Rodríguez-Ezpeleta, N. & Embley, T. M. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS ONE 7, e30520 (2012).
Viale, A. M. & Arakaki, A. K. The chaperone connection to the origins of the eukaryotic organelles. FEBS Lett. 341, 146–151 (1994).
Castelli, M. et al. Deianiraea, an extracellular bacterium associated with the ciliate Paramecium, suggests an alternative scenario for the evolution of Rickettsiales. ISME J. 13, 2280–2294 (2019).
Brindefalk, B., Ettema, T. J., Viklund, J., Thollesson, M. & Andersson, S. G. A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS ONE 6, e24457 (2011).
Thrash, J. C. et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1, 13 (2011).
Georgiades, K., Madoui, M. A., Le, P., Robert, C. & Raoult, D. Phylogenomic analysis of Odyssella thessalonicensis fortifies the common origin of Rickettsiales, Pelagibacter ubique and Reclimonas americana mitochondrion. PLoS ONE 6, e24857 (2011).
Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T. J. G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018).
Gray, M. W., Burger, G. & Lang, B. F. Mitochondrial evolution. Science 283, 1476–1481 (1999).
Gawryluk, R. M. R. Evolutionary biology: A new home for the powerhouse? Curr. Biol. 28, R798–R800 (2018).
Blanquart, S. & Lartillot, N. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol. Biol. Evol. 23, 2058–2071 (2006).
Muñoz-Gómez, S. A. et al. An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. eLife 8, e42535 (2019).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Jermiin, L. S., Jayaswal, V., Ababneh, F. M. & Robinson, J. Identifying optimal models of evolution. Methods Mol. Biol. 1525, 379–420 (2017).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615 (2013).
Kannan, S., Rogozin, I. B. & Koonin, E. V. MitoCOGs: clusters of orthologous genes from mitochondria and implications for the evolution of eukaryotes. BMC Evol. Biol. 14, 237 (2014).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Criscuolo, A. & Gribaldo, S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Fan, L. et al. Mitochondria and Alphaproteobacteria phylogenetic study alignments and tree files. figshare https://doi.org/10.6084/m9.figshare.12347216 (2020).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (91851210, 91951120, 41530105 and 81774152), the European Research Council (ERC 666053), the Shenzhen Key Laboratory of Marine Archaea Geo-Omics, Southern University of Science and Technology (ZDSYS201802081843490), the Shenzhen Science and Technology Innovation Commission (JCYJ20180305123458107), the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (K19313901) and the VW foundation (93 046). Computation in this study was supported by the Centre for Computational Science and Engineering at the Southern University of Science and Technology.
Author information
Authors and Affiliations
Contributions
L.F., W.F.M. and R.Z. conceived this study. L.F., D.W., V.G., J.X., Y.X. and S.G. were involved in data analysis. L.F., V.G., C.Z., W.F.M. and R.Z. interpreted the results and drafted the manuscript. All authors participated in the critical revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, references, Notes 1–9 and Figs. 1–60.
Supplementary Table
Supplementary Tables 1–7.
Supplementary Software
The script of the Bowker’s test score-based site-exclusion method.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, L., Wu, D., Goremykin, V. et al. Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria. Nat Ecol Evol 4, 1213–1219 (2020). https://doi.org/10.1038/s41559-020-1239-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-020-1239-x
This article is cited by
-
Mechanism and role of mitophagy in the development of severe infection
Cell Death Discovery (2024)
-
Host association and intracellularity evolved multiple times independently in the Rickettsiales
Nature Communications (2024)
-
Complete mitogenome assembly of Selenicereus monacanthus revealed its molecular features, genome evolution, and phylogenetic implications
BMC Plant Biology (2023)
-
Phylogenetic affiliation of mitochondria with Alpha-II and Rickettsiales is an artefact
Nature Ecology & Evolution (2022)
-
Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria
Nature Ecology & Evolution (2022)