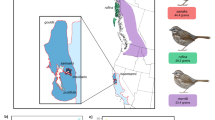
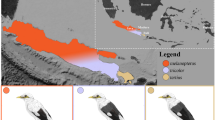
Abstract
To cope with climate change, species may shift their distributions or adapt in situ to changing environmental conditions. However, clear examples of genetic changes via adaptation are limited. We explore evolutionary responses to climate change in the endangered southwestern willow flycatcher (Empidonax traillii extimus) through whole-genome comparisons between historical specimens, collected from 1888 to 1909 near San Diego, California, United States, and contemporary individuals from across the breeding range. Genomic analyses revealed that introgression into San Diego increased adaptive potential over time and shifted genome-wide population structure towards that of neighbouring populations. In contrast, loci linked to climate (dew point temperature and precipitation) shifted away from neighbouring populations and in a direction consistent with adaptation to climate change in southern California. This research highlights the role of admixture in facilitating adaptive shifts through its impact on genome-wide genetic variation and represents one of the few studies to document climate adaptation in a wild population.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data generated for this study are available in the NCBI Sequence Read Archive (BioProject PRJNA957938) and Dryad Digital Repository (https://datadryad.org/stash/share/qlafA02RwIPjoptjefUaKbrMszEsMJWP8a_H9-e24MY)68.
Code availability
Code associated with this study can be accessed on GitHub (https://github.com/sturbek/WIFL-Climate-Adaptation)69.
References
Hoffmann, A. A. & Sgrò, C. M. Climate change and evolutionary adaptation. Nature 470, 479–485 (2011).
Razgour, O. et al. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc. Natl Acad. Sci. USA 116, 10418–10423 (2019).
Catullo, R. A., Llewelyn, J., Phillips, B. L. & Moritz, C. C. The potential for rapid evolution under anthropogenic climate change. Curr. Biol. 29, R996–R1007 (2019).
Merilä, J. & Hendry, A. P. Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol. Appl 7, 1–14 (2014).
Bi, K. et al. Temporal genomic contrasts reveal rapid evolutionary responses in an alpine mammal during recent climate change. PLoS Genet. 15, e1008119 (2019).
Geerts, A. N. et al. Rapid evolution of thermal tolerance in the water flea Daphnia. Nat. Clim. Change 5, 665–668 (2015).
Umina, P. A., Weeks, A. R., Kearney, M. R., McKechnie, S. W. & Hoffmann, A. A. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691–693 (2005).
Kardos, M. et al. The crucial role of genome-wide genetic variation in conservation. Proc. Natl Acad. Sci. USA 118, e2104642118 (2021).
Eizaguirre, C. & Baltazar-Soares, M. Evolutionary conservation—evaluating the adaptive potential of species. Evol. Appl. 7, 963–967 (2014).
Aitken, S. N. & Whitlock, M. C. Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 44, 367–388 (2013).
Bell, D. A. et al. The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol. 34, 1070–1079 (2019).
Chan, W. Y., Hoffmann, A. A. & van Oppen, M. J. H. Hybridization as a conservation management tool. Conserv. Lett. 12, e12652 (2019).
Billerman, S. M. & Walsh, J. Historical DNA as a tool to address key questions in avian biology and evolution: a review of methods, challenges, applications, and future directions. Mol. Ecol. Resour. 19, 1115–1130 (2019).
Sogge, M. K., Marshall, R. M., Sferra, S. J. & Tibbitts, T. J. A Southwestern Willow Flycatcher Natural History Summary and Survey Protocol. Technical Report NPS/NAUCPRS/NRTR-97/12 (USGS, 1997).
Diffenbaugh, N. S., Giorgi, F. & Pal, J. S. Climate change hotspots in the United States. Geophys. Res. Lett. 35, L16709 (2008).
Ruegg, K. et al. Ecological genomics predicts climate vulnerability in an endangered southwestern songbird. Ecol. Lett. 21, 1085–1096 (2018).
Southwestern Willow Flycatcher (Empidonax traillii extimus). 5-Year Review: Summary and Evaluation (US Fish and Wildlife Service, 2014).
Unitt, P. Empidonax traillii extimus: an endangered subspecies. West. Birds 18, 137–162 (1987).
Indicators of Climate Change in California 4th edn (California Environmental Protection Agency, OEHHA, 2022).
Ruegg, K. et al. Linking climate niches across seasons to assess population vulnerability in a migratory bird. Glob. Change Biol. 27, 3519–3531 (2021).
Hamilton, J. A. & Miller, J. M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 30, 33–41 (2016).
Whiteley, A. R., Fitzpatrick, S. W., Funk, W. C. & Tallmon, D. A. Genetic rescue to the rescue. Trends Ecol. Evol. 30, 42–49 (2015).
DeWoody, J. A., Harder, A. M., Mathur, S. & Willoughby, J. R. The long-standing significance of genetic diversity in conservation. Mol. Ecol. 30, 4147–4154 (2021).
Lavergne, S. & Molofsky, J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl Acad. Sci. USA 104, 3883–3888 (2007).
Nolte, A. W., Freyhof, J., Stemshorn, K. C. & Tautz, D. An invasive lineage of sculpins, Cottus sp. (Pisces, Teleostei) in the Rhine with new habitat adaptations has originated from hybridization between old phylogeographic groups. Proc. R. Soc. B 272, 2379–2387 (2005).
Racimo, F., Sankararaman, S., Nielsen, R. & Huerta-Sánchez, E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359–371 (2015).
Becker, M. et al. Hybridization may facilitate in situ survival of endemic species through periods of climate change. Nat. Clim. Change 3, 1039–1043 (2013).
Bay, R. A. et al. Genomic signals of selection predict climate-driven population declines in a migratory bird. Science 359, 83–86 (2018).
Fitzpatrick, M. C. & Keller, S. R. Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 18, 1–16 (2015).
Kus, B. E., Beck, P. P. & Wells, J. M. Southwestern willow flycatcher populations in California: distribution, abundance, and potential for conservation. Stud. Avian Biol. 26, 12–21 (2003).
Bird Species Distribution Maps of the World (BirdLife International & NatureServe, 2012).
Paxton, E. H. Molecular Genetic Structuring and Demographic History of the Willow Flycatcher, Empidonax traillii (Northern Arizona Univ., 2000).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Picard Toolkit (Broad Institute, 2018); http://broadinstitute.github.io/picard/
Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011).
Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F. & Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013).
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: analysis of next generation sequencing data. BMC Bioinf. 15, 356 (2014).
Li, H. Improving SNP discovery by base alignment quality. Bioinformatics 27, 1157–1158 (2011).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Raxworthy, C. J. & Smith, B. T. Mining museums for historical DNA: advances and challenges in museomics. Trends Ecol. Evol. 36, 1049–1060 (2021).
Meisner, J. & Albrechtsen, A. Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731 (2018).
Joseph, T. A. & Pe’er, I. Inference of population structure from time-series genotype data. Am. J. Hum. Genet. 105, 317–333 (2019).
Grabherr, M. G. et al. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics 26, 1145–1151 (2010).
Turner, S. D. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. J. Open Source Softw. 3, 731 (2018).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020).
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).
Petr, M., Vernot, B. & Kelso, J. admixr—R package for reproducible analyses using ADMIXTOOLS. Bioinformatics 35, 3194–3195 (2019).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Do, C. et al. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 14, 209–214 (2014).
Biscarini, F., Cozzi, P., Gaspa, G. & Marras, G. detectRUNS: Detect runs of homozygosity and runs of heterozygosity in diploid genomes. R package version 0.9.6 (2019).
Goudet, J. hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186 (2005).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Ellis, N., Smith, S. J. & Pitcher, C. R. Gradient forests: calculating importance gradients on physical predictors. Ecology 93, 156–168 (2012).
Forester, B. R., Lasky, J. R., Wagner, H. H. & Urban, D. L. Comparing methods for detecting multilocus adaptation with multivariate genotype–environment associations. Mol. Ecol. 27, 2215–2233 (2018).
Browning, B. L., Zhou, Y. & Browning, S. R. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103, 338–348 (2018).
Oksanen, J. et al. vegan: Community ecology package. R package version 2.6-4 (2022).
Forester, B. R., Jones, M. R., Joost, S., Landguth, E. L. & Lasky, J. R. Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol. Ecol. 25, 104–120 (2016).
Frichot, E. & François, O. LEA: an R package for landscape and ecological association studies. Methods Ecol. Evol. 6, 925–929 (2015).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Backström, N., Qvarnström, A., Gustafsson, L. & Ellegren, H. Levels of linkage disequilibrium in a wild bird population. Biol. Lett. 2, 435–438 (2006).
Vitorino Carvalho, A. et al. Embryonic thermal manipulation impacts the postnatal transcriptome response of heat-challenged Japanese quails. BMC Genom. 22, 488 (2021).
Kumar, H. et al. Transcriptome of chicken liver tissues reveals the candidate genes and pathways responsible for adaptation into two different climatic conditions. Animals 9, 1076 (2019).
Zhang, J., Schmidt, C. J. & Lamont, S. J. Transcriptome analysis reveals potential mechanisms underlying differential heart development in fast- and slow-growing broilers under heat stress. BMC Genom. 18, 295 (2017).
Wang, Y. et al. Liver transcriptome responses to heat stress and Newcastle disease virus infection in genetically distinct chicken inbred lines. Genes 11, 1067 (2020).
Turbek, S. T. et al. Data for ‘Historical DNA reveals climate adaptation in an endangered songbird’. Dryad https://datadryad.org/stash/share/qlafA02RwIPjoptjefUaKbrMszEsMJWP8a_H9-e24MY (2023).
Turbek, S. T. et al. Code for ‘Historical DNA reveals climate adaptation in an endangered songbird’. GitHub https://github.com/sturbek/WIFL-Climate-Adaptation (2023).
Acknowledgements
This work was supported by a National Science Foundation (NSF) Postdoctoral Research Fellowship in Biology (2208881) to S.T. and an NSF CAREER grant (1942313) to K.R. We thank B. Forester for contributing funding for sequencing. We thank all of the individuals who contributed genetic samples, including T. Kita, B. Keith, R. Taylor and S. Birks. The following museums generously provided historical samples for the analyses presented in this study: the University of California Berkeley Museum of Vertebrate Zoology, California Academy of Sciences, Los Angeles County Natural History Museum, American Museum of Natural History, Buffalo Museum of Science, Charles R. Conner Museum, Cornell University Museum of Vertebrates, University of Kansas Biodiversity Institute & Natural History Museum, New York State Museum, Royal Ontario Museum, University of Colorado Museum of Natural History and the University of Michigan Museum of Zoology. All samples were collected under federal, state and IACUC permits necessary for the research. Any use of trade, firm or product names is for descriptive purposes only and does not imply endorsement by the US government.
Author information
Authors and Affiliations
Contributions
S.T. and K.R. designed the study. B.K., M.W. and E.P. contributed samples for genomic analysis. R.B. and K.R. obtained museum samples. C.R. and C.G. generated the genomic data. S.T. and C.B. analysed the genomic data. K.R. funded the study. K.R. and T.S. provided logistical support. S.T. wrote the paper with edits from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Climate Change thanks Fumin Lei, Orly Razgour and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Genomic differentiation between historical and contemporary willow flycatchers in San Diego.
Manhattan plot showing mean FST, averaged over 25-kb windows, between historical (n = 17) and contemporary (n = 18) individuals breeding in San Diego. Colours (black and grey) alternate between scaffolds in the willow flycatcher reference genome.
Extended Data Fig. 2 Results from the gradient forest analysis of 500,000 loci and 11 contemporary populations across the breeding range of the southwestern willow flycatcher.
(A) PCA of the top three climate variables associated with genomic variation. The arrows indicate the loadings of the top-ranking environmental variables. Colours are based on the predicted genomic composition from the top climate variables at 100,000 random points. (B) Gradient forest-transformed environmental variables from the PCA mapped across the breeding range of the southwestern willow flycatcher. Sampling locations are indicated with black circles.
Extended Data Fig. 3 Redundancy analysis (RDA) showing the relationship between genomic variation and top-ranking climate variables for 221 contemporary willow flycatchers breeding across the United States.
The loadings on (A) axes one and two and (B) axes one and three for monthly mean dew point temperature (tdmean), monthly precipitation (precip), and monthly maximum temperature (tmax) are shown. Points are coloured by sampling location and the grey cloud in the center of each plot shows the 128,147 SNPs included in the analysis. The four subspecies of willow flycatcher are shown as different shapes.
Extended Data Fig. 4 Redundancy analysis (RDA) showing the candidate SNPs associated with top-ranking climate variables for 221 contemporary willow flycatchers breeding across the United States.
The loadings of 128,147 SNPs on (A) axes one and two and (B) axes one and three and their relationship with monthly mean dew point temperature (tdmean), monthly precipitation (precip), and monthly maximum temperature (tmax) are shown. Candidate SNPs (with loadings greater than three standard deviations from the mean) are coloured according to their associations, while non-candidate SNPs are shown in light grey.
Extended Data Fig. 5 Shifts in patterns of genomic differentiation over time in willow flycatchers breeding across the United States at climate-linked SNPs.
Principal component analysis (PCA) of loci associated with (A) monthly mean dew point temperature (n = 104), (B) monthly precipitation (n = 72), and (C) monthly maximum temperature (n = 56) across 238 contemporary and historical samples of the willow flycatcher. Details as in Fig. 1. Contemporary samples of the four subspecies are shown as squares, circles, diamonds, and triangles, while upside-down triangles indicate historical samples.
Extended Data Fig. 6 Allele frequency shifts within San Diego at climate-linked loci located near genes associated with avian thermal tolerance and bill morphology.
Number of loci linked to (A) monthly mean dew point temperature (n = 20), (B) monthly precipitation (n = 10), and (C) monthly maximum temperature (n = 7) in willow flycatchers breeding in San Diego that shifted in a direction consistent with climate adaptation. The p-values show the results for one-tailed binomial tests examining whether the observed proportion of loci that shifted in the expected direction (that is, consistent with climate adaptation) was significantly greater than expected by chance.
Supplementary information
Supplementary Information
Supplementary Methods, Tables 1–5 and Figs. 1–15.
Supplementary Table
Fluidigm primer information (5’ to 3’) for the 96 SNP markers included in the SNP genotyping analyses. ASP1, the SNP allele detected with allele-specific primer 1; ASP2, the SNP allele detected with allele-specific primer 2; SNP_SEQ, the sequence of the amplified fragment containing the SNP; ASP1_SEQ, the sequence of allele-specific primer 1; ASP2_SEQ, the sequence of allele-specific primer 2; LSP_SEQ, the sequence of locus-specific reverse primer; STA_SEQ, the sequence of forward primer for specific target amplification; AMP_GC, the proportional GC content.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Turbek, S.P., Bossu, C., Rayne, C. et al. Historical DNA reveals climate adaptation in an endangered songbird. Nat. Clim. Chang. 13, 735–741 (2023). https://doi.org/10.1038/s41558-023-01696-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41558-023-01696-3
This article is cited by
-
Historical DNA reveals climate adaptation in an endangered songbird
Nature Climate Change (2023)
-
Museum specimens uncover the past, present and future
Nature Climate Change (2023)