Abstract
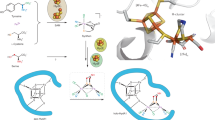
Understanding the chemical bonding in the catalytic cofactor of the Mo nitrogenase (FeMo-co) is foundational for building a mechanistic picture of biological nitrogen fixation. A persistent obstacle towards this goal has been that the 57Fe-based spectroscopic data—although rich with information—combines responses from all seven Fe sites, and it has therefore not been possible to map individual spectroscopic responses to specific sites in the three-dimensional structure. Here we have addressed this challenge by incorporating 57Fe into a single site of FeMo-co. Spectroscopic analysis of the resting state informed on the local electronic structure of the terminal Fe1 site, including its oxidation state and spin orientation, and, in turn, on the spin-coupling scheme for the entire cluster. The oxidized resting state and the first intermediate in nitrogen fixation were also characterized, and comparisons with the resting state provided molecular-level insights into the redox chemistry of FeMo-co.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data supporting the findings of this work are available within the article and its Supplementary Information. Data supporting the current study are also available from the corresponding author upon request. PDB 3U7Q was used in the preparation of Fig. 1.
References
Raymond, J., Siefert, J. L., Staples, C. R. & Blankenship, R. E. The natural history of nitrogen fixation. Mol. Biol. Evol. 21, 541–554 (2004).
Canfield, D. E., Glazer, A. N. & Falkowski, P. G. The evolution and future of Earth’s nitrogen cycle. Science 330, 192–196 (2010).
Hoffman, B. M., Lukoyanov, D., Yang, Z.-Y., Dean, D. R. & Seefeldt, L. C. Mechanism of nitrogen fixation by nitrogenase: the next stage. Chem. Rev. 114, 4041–4062 (2014).
Winter, H. C. & Burris, R. H. Nitrogenase. Annu. Rev. Biochem. 45, 409–426 (1976).
Burgess, B. K. & Lowe, D. J. Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3012 (1996).
Van Stappen, C. et al. The spectroscopy of nitrogenases. Chem. Rev. 120, 5005–5081 (2020).
Einsle, O. & Rees, D. C. Structural enzymology of nitrogenase enzymes. Chem. Rev. 120, 4969–5004 (2020).
Seefeldt, L. C. et al. Reduction of substrates by nitrogenases. Chem. Rev. 120, 5082–5106 (2020).
Spatzal, T. et al. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940 (2011).
Lovell, T., Li, J., Liu, T., Case, D. A. & Noodleman, L. FeMo cofactor of nitrogenase: a density functional study of states MN, MOX, MR, and MI. J. Am. Chem. Soc. 123, 12392–12410 (2001).
Harris, T. V. & Szilagyi, R. K. Comparative assessment of the composition and charge state of nitrogenase FeMo-cofactor. Inorg. Chem. 50, 4811–4824 (2011).
Siegbahn, P. E. M. Model calculations suggest that the central carbon in the FeMo-cofactor of nitrogenase becomes protonated in the process of nitrogen fixation. J. Am. Chem. Soc. 138, 10485–10495 (2016).
Bjornsson, R., Neese, F. & DeBeer, S. Revisiting the Mössbauer isomer shifts of the FeMoco cluster of nitrogenase and the cofactor charge. Inorg. Chem. 56, 1470–1477 (2017).
Benediktsson, B. & Bjornsson, R. QM/MM study of the nitrogenase MoFe protein resting state: broken-symmetry states, protonation states, and QM region convergence in the FeMoco active site. Inorg. Chem. 56, 13417–13429 (2017).
Raugei, S., Seefeldt, L. C. & Hoffman, B. M. Critical computational analysis illuminates the reductive-elimination mechanism that activates nitrogenase for N2 reduction. Proc. Natl Acad. Sci. USA 115, E10521 (2018).
Li, Z., Li, J., Dattani, N. S., Umrigar, C. J. & Chan, G. K.-L. The electronic complexity of the ground-state of the FeMo cofactor of nitrogenase as relevant to quantum simulations. J. Chem. Phys. 150, 024302 (2019).
Cramer, S. P., Hodgson, K. O., Gillum, W. O. & Mortenson, L. E. The molybdenum site of nitrogenase. Preliminary structural evidence from X-ray absorption spectroscopy. J. Am. Chem. Soc. 100, 3398–3407 (1978).
Cramer, S. P. et al. The molybdenum site of nitrogenase. 2. A comparative study of molybdenum–iron proteins and the iron–molybdenum cofactor by X-ray absorption spectroscopy. J. Am. Chem. Soc. 100, 3814–3819 (1978).
Conradson, S. D. et al. Structural insights from the molybdenum K-edge X-ray absorption near edge structure of the iron–molybdenum protein of nitrogenase and its iron–molybdenum cofactor by comparison with synthetic iron–molybdenum–sulfur clusters. J. Am. Chem. Soc. 107, 7935–7940 (1985).
Venters, R. A. et al. ENDOR of the resting state of nitrogenase molybdenum–iron proteins from Azotobacter vinelandii, Klebsiella pneumoniae, and Clostridium pasteurianum. Proton, iron-57, molybdenum-95, and sulfur-33 studies. J. Am. Chem. Soc. 108, 3487–3498 (1986).
Lukoyanov, D., Yang, Z.-Y., Dean, D. R., Seefeldt, L. C. & Hoffman, B. M. Is Mo involved in hydride binding by the four-electron reduced (E4) intermediate of the nitrogenase MoFe protein? J. Am. Chem. Soc. 132, 2526–2527 (2010).
Bjornsson, R. et al. Identification of a spin-coupled Mo(III) in the nitrogenase iron–molybdenum cofactor. Chem. Sci. 5, 3096–3103 (2014).
Van Stappen, C. et al. Spectroscopic description of the E1 state of Mo nitrogenase based on Mo and Fe X-ray absorption and Mössbauer studies. Inorg. Chem. 58, 12365–12376 (2019).
True, A. E., Nelson, M. J., Venters, R. A., Orme-Johnson, W. H. & Hoffman, B. M. Iron-57 hyperfine coupling tensors of the FeMo cluster in Azotobacter vinelandii MoFe protein: determination by polycrystalline ENDOR spectroscopy. J. Am. Chem. Soc. 110, 1935–1943 (1988).
Yoo, S. J., Angove, H. C., Papaefthymiou, V., Burgess, B. K. & Münck, E. Mössbauer study of the MoFe protein of nitrogenase from Azotobacter vinelandii using selective 57Fe enrichment of the M-centers. J. Am. Chem. Soc. 122, 4926–4936 (2000).
Lukoyanov, D. A. et al. Electron redistribution within the nitrogenase active site FeMo-cofactor during reductive elimination of H2 to achieve N≡N triple-bond activation. J. Am. Chem. Soc. 142, 21679–21690 (2020).
Münck, E. et al. Nitrogenase. VIII. Mössbauer and EPR spectroscopy. The MoFe protein component from Azotobacter vinelandii OP. Biochim. Biophys. Acta Protein Struct. 400, 32–53 (1975).
Spatzal, T. et al. Nitrogenase FeMoco investigated by spatially resolved anomalous dispersion refinement. Nat. Commun. 7, 10902 (2016).
Srisantitham, S., Badding, E. D. & Suess, D. L. M. Postbiosynthetic modification of a precursor to the nitrogenase iron–molybdenum cofactor. Proc. Natl Acad. Sci. USA 118, e2015361118 (2021).
Ribbe, M. W., Hu, Y., Hodgson, K. O. & Hedman, B. Biosynthesis of nitrogenase metalloclusters. Chem. Rev. 114, 4063–4080 (2014).
Burén, S., Jiménez-Vicente, E., Echavarri-Erasun, C. & Rubio, L. M. Biosynthesis of nitrogenase cofactors. Chem. Rev. 120, 4921–4968 (2020).
Shah, V. K. & Brill, W. J. Isolation of an iron–molybdenum cofactor from nitrogenase. Proc. Natl Acad. Sci. USA 74, 3249–3253 (1977).
Rawlings, J. et al. Novel metal cluster in the iron–molybdenum cofactor of nitrogenase. Spectroscopic evidence. J. Biol. Chem. 253, 1001–1004 (1978).
Yang, S. S. et al. Iron–molybdenum cofactor from nitrogenase. Modified extraction methods as probes for composition. J. Biol. Chem. 257, 8042–8048 (1982).
Mouesca, J. M., Noodleman, L., Case, D. A. & Lamotte, B. Spin densities and spin coupling in iron–sulfur clusters: a new analysis of hyperfine coupling constants. Inorg. Chem. 34, 4347–4359 (1995).
Pandelia, M.-E., Lanz, N. D., Booker, S. J. & Krebs, C. Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta Mol. Cell Res. 1853, 1395–1405 (2015).
Papaefthymiou, V., Girerd, J. J., Moura, I., Moura, J. J. G. & Muenck, E. Moessbauer study of D. gigas ferredoxin II and spin-coupling model for Fe3S4 cluster with valence delocalization. J. Am. Chem. Soc. 109, 4703–4710 (1987).
Noodleman, L. Exchange coupling and resonance delocalization in reduced iron–sulfur [Fe4S4]+ and iron–selenium [Fe4Se4]+ clusters. 1. Basic theory of spin-state energies and EPR and hyperfine properties. Inorg. Chem. 30, 246–256 (1991).
Venkateswara Rao, P. & Holm, R. H. Synthetic analogues of the active sites of iron–sulfur proteins. Chem. Rev. 104, 527–560 (2004).
Solomon, E. I., Gorelsky, S. I. & Dey, A. Metal–thiolate bonds in bioinorganic chemistry. J. Comput. Chem. 27, 1415–1428 (2006).
Lukoyanov, D. et al. Testing if the interstitial atom, X, of the nitrogenase molybdenum–iron cofactor is N or C: ENDOR, ESEEM, and DFT studies of the S = 3/2 resting state in multiple environments. Inorg. Chem. 46, 11437–11449 (2007).
Lukoyanov, D. A. et al. 13C ENDOR characterization of the central carbon within the nitrogenase catalytic cofactor indicates that the CFe6 core is a stabilizing ‘heart of steel’. J. Am. Chem. Soc. 144, 18315–18328 (2022).
Pérez-González, A. et al. Exploring the role of the central carbide of the nitrogenase active-site FeMo-cofactor through targeted 13C labeling and ENDOR spectroscopy. J. Am. Chem. Soc. 143, 9183–9190 (2021).
Zimmermann, R. et al. Nitrogenase X: Mössbauer and EPR studies on reversibly oxidized MoFe protein from Azotobacter vinelandii OP. Nature of the iron centers. Biochim. Biophys. Acta Protein Struct. 537, 185–207 (1978).
Johnson, M. K., Thomson, A. J., Robinson, A. E. & Smith, B. E. Characterization of the paramagnetic centres of the molybdenum–iron protein of nitrogenase from Klebsiella pneumoniae using low temperature magnetic circular dichroism spectroscopy. Biochim. Biophys. Acta Protein Struct. 671, 61–70 (1981).
Lindahl, P. A., Papaefthymiou, V., Orme-Johnson, W. H. & Münck, E. Mössbauer studies of solid thionin-oxidized MoFe protein of nitrogenase. J. Biol. Chem. 263, 19412–19418 (1988).
Smith, B. E. & Lang, G. Mössbauer spectroscopy of the nitrogenase proteins from Klebsiella pneumoniae. Structural assignments and mechanistic conclusions. Biochem. J 137, 169–180 (1974).
Surerus, K. K. et al. Moessbauer and integer-spin EPR of the oxidized P-clusters of nitrogenase: POX is a non-Kramers system with a nearly degenerate ground doublet. J. Am. Chem. Soc. 114, 8579–8590 (1992).
Spatzal, T., Perez, K. A., Einsle, O., Howard, J. B. & Rees, D. C. Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase. Science 345, 1620–1623 (2014).
Spatzal, T., Perez, K. A., Howard, J. B. & Rees, D. C. Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron–molybdenum cofactor. eLife 4, e11620 (2015).
Kang, W., Lee, C. C., Jasniewski, A. J., Ribbe, M. W. & Hu, Y. Structural evidence for a dynamic metallocofactor during N2 reduction by Mo-nitrogenase. Science 368, 1381–1385 (2020).
Lee, C. C. et al. Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor. Nat. Catal. 5, 443–454 (2022).
Van Stappen, C., Thorhallsson, A. T., Decamps, L., Bjornsson, R. & DeBeer, S. Resolving the structure of the E1 state of Mo nitrogenase through Mo and Fe K-edge EXAFS and QM/MM calculations. Chem. Sci. 10, 9807–9821 (2019).
Christiansen, J., Tittsworth, R. C., Hales, B. J. & Cramer, S. P. Fe and Mo EXAFS of Azotobacter vinelandii nitrogenase in partially oxidized and singly reduced forms. J. Am. Chem. Soc. 117, 10017–10024 (1995).
Lukoyanov, D. A. et al. The one-electron reduced active-site FeFe-cofactor of Fe-nitrogenase contains a hydride bound to a formally oxidized metal-ion core. Inorg. Chem. 61, 5459–5464 (2022).
Cao, L., Caldararu, O. & Ryde, U. Protonation and reduction of the FeMo cluster in nitrogenase studied by quantum mechanics/molecular mechanics (QM/MM) calculations. J. Comput. Chem. 14, 6653–6678 (2018).
Lee, C.-C., Ribbe, M. W. & Hu, Y. in Metalloproteins: Methods and Protocols (ed. Hu, Y.) 111–124 (Springer, 2019).
Burgess, B. K., Jacobs, D. B. & Stiefel, E. I. Large-scale purification of high activity Azotobacter vinelandII nitrogenase. Biochim. Biophys. Acta Enzymol. 614, 196–209 (1980).
McLean, P. A., Papaefthymiou, V., Orme-Johnson, W. H. & Münck, E. Isotopic hybrids of nitrogenase. Mössbauer study of MoFe protein with selective 57Fe enrichment of the P-cluster. J. Biol. Chem. 262, 12900–12903 (1987).
Christiansen, J., Goodwin, P. J., Lanzilotta, W. N., Seefeldt, L. C. & Dean, D. R. Catalytic and biophysical properties of a nitrogenase apo-MoFe protein produced by a nifB-deletion mutant of Azotobacter vinelandii. Biochemistry 37, 12611–12623 (1998).
Harris, D. F., Yang, Z.-Y., Dean, D. R., Seefeldt, L. C. & Hoffman, B. M. Kinetic understanding of N2 reduction versus H2 evolution at the E4(4H) Janus state in the three nitrogenases. Biochemistry 57, 5706–5714 (2018).
Davoust, C. E., Doan, P. E. & Hoffman, B. M. Q-band pulsed electron spin-echo spectrometer and its application to ENDOR and ESEEM. J. Magn. Reson. A 119, 38–44 (1996).
Acknowledgements
We acknowledge the National Institutes of Health (GM141203 to D.L.M.S. and GM111097 to B.M.H.), the MIT Research Support Committee (to D.L.M.S.) and the National Science Foundation (MCB-1908587 to B.M.H.) for funding. We thank N. Thompson for helpful discussions as well as D. Dean and V. Cash for providing the Azotobacter vinelandii strains used in this work. Support for the ICP-MS instrument was provided by a core centre grant (P30-ES002109) from the National Institute of Environmental Health Sciences, NIH.
Author information
Authors and Affiliations
Contributions
E.D.B., S.S. and D.A.L. performed the experiments. All authors contributed to the design of the study, data analysis and paper writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Lou Noodleman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20, Tables 1–22, Discussion and References.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Badding, E.D., Srisantitham, S., Lukoyanov, D.A. et al. Connecting the geometric and electronic structures of the nitrogenase iron–molybdenum cofactor through site-selective 57Fe labelling. Nat. Chem. 15, 658–665 (2023). https://doi.org/10.1038/s41557-023-01154-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-023-01154-9