Abstract
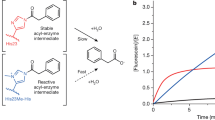
The combination of computational design and directed evolution could offer a general strategy to create enzymes with new functions. So far, this approach has delivered enzymes for a handful of model reactions. Here we show that new catalytic mechanisms can be engineered into proteins to accelerate more challenging chemical transformations. Evolutionary optimization of a primitive design afforded an efficient and enantioselective enzyme (BH32.14) for the Morita–Baylis–Hillman (MBH) reaction. BH32.14 is suitable for preparative-scale transformations, accepts a broad range of aldehyde and enone coupling partners and is able to promote selective monofunctionalizations of dialdehydes. Crystallographic, biochemical and computational studies reveal that BH32.14 operates via a sophisticated catalytic mechanism comprising a His23 nucleophile paired with a judiciously positioned Arg124. This catalytic arginine shuttles between conformational states to stabilize multiple oxyanion intermediates and serves as a genetically encoded surrogate of privileged bidentate hydrogen-bonding catalysts (for example, thioureas). This study demonstrates that elaborate catalytic devices can be built from scratch to promote demanding multi-step processes not observed in nature.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 6Z1K, 7O1D and 6Z1L. Data supporting the findings of this study are available within the paper and its Supplementary Information. Source data are provided with this paper.
References
Hilvert, D. Design of protein catalysts. Annu. Rev. Biochem. 82, 447–470 (2013).
Röthlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008).
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science 329, 309–313 (2010).
Eiben, C. B. et al. Increased Diels–alderase activity through backbone remodelling guided by Foldit players. Nat. Biotechnol. 30, 190–192 (2012).
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013).
Obexer, R. et al. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat. Chem. 9, 50–56 (2017).
Gouverneur, V. E. et al. Control of the exo and endo pathways of the Diels-Alder reaction by antibody catalysis. Science 262, 204–208 (1993).
Stewart, J. D. & Benkovic, S. J. Transition-state stabilization as a measure of the efficiency of antibody catalysis. Nature 375, 388–391 (1995).
Barbas, C. F. III et al. Immune versus natural selection: antibody aldolases with enzymic rates but broader scope. Science 278, 2085–2092 (1997).
Basavaiah, D., Rao, A. J. & Satyanarayana, T. Recent advances in the Baylis-Hillman reaction and applications. Chem. Rev. 103, 811–892 (2003).
Basavaiah, D., Reddy, B. S. & Badsara, S. S. Recent contributions from the Baylis-Hillman reaction to organic chemistry. Chem. Rev. 9, 5447–5674 (2010).
Wei, Y. & Shi, M. Recent advances in organocatalytic asymmetric Morita-Baylis-Hillman/aza-Morita-Baylis-Hillman reactions. Chem. Rev. 113, 6659–6690 (2013).
Metrano, A. J. et al. Asymmetric catalysis mediated by synthetic peptides, version 2.0: expansion of scope and mechanisms. Chem. Rev. 120, 11479–11615 (2020).
Shi, M. & Liu, X.-G. Asymmetric Morita-Baylis-Hillman reaction of arylaldehydes with 2-cyclohexen-1-one and 2-cyclopenten-1-one catalyzed by chiral bis(thio)urea and DABCO. Org. Lett. 10, 1043–1046 (2008).
Nakayama, Y., Gotanda, T. & Ito, K. Asymmetric Morita-Baylis-Hillman reactions of 2-cyclohexen-1-one catalyzed by chiral biaryl-based bis(thiourea) organocatalysts. Tetrahedron Lett. 52, 6234–6237 (2011).
Islam, Z., Strutzenberg, T. S., Gurevic, I. & Kohen, A. Concerted versus stepwise mechanism in thymidylate synthase. J. Am. Chem. Soc. 136, 9850–9853 (2014).
Reetz, M. T., Mondière, R. & Carballeira, J. D. Enzyme promiscuity: first protein-catalyzed Morita-Baylis-Hillman reaction. Tetrahedron Lett. 48, 1679–1681 (2007).
López-Iglesias, M., Busto, E., Gotor, V. & Gotor-Fernández, V. Use of protease from Bacillus licheniformis as promiscuous catalyst for organic synthesis: applications in C-C and C-N bond formation reactions. Adv. Synth. Catal. 353, 2345–2353 (2011).
Joshi, P. N., Purushottam, L., Das, N. K., Mukherjee, S. & Rai, V. Protein self-assembly induces promiscuous nucleophilic biocatalysis in Morita-Baylis-Hillman (MBH) reaction. RSC Adv. 6, 208–211 (2016).
Bjelic, S. et al. Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction. ACS Chem. Biol. 8, 749–757 (2013).
Bloom, J. D., Labthavikul, S. T., Otey, C. R. & Arnold, F. H. Protein stability promotes evolvability. Proc. Natl Acad. Sci. USA 103, 5869–5874 (2006).
Tokuriki, N. & Tawfik, D. S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19, 596–604 (2009).
Aggarwal, V. K., Emme, I. & Fulford, S. Y. Correlation between pKa and reactivity of quinuclidine-based catalysts in the Baylis-Hillman reaction: discovery of quinuclidine as optimum catalyst leading to substantial enhancement of scope. J. Org. Chem. 3, 692–700 (2003).
Karur, S., Hardin, J., Headley, A. & Li, G. A novel approach to Morita-Baylis-Hillman (MBH) lactones via the Lewis acid-promoted couplings of α,β-unsaturated lactone with aldehydes. Tetrahedron Lett. 44, 2991–2994 (2003).
Phillips, M. A., Fletterick, R. & Rutter, W. J. Arginine 127 stabilizes the transition state in carboxypeptidase. J. Biol. Chem. 265, 20692–20698 (1990).
Studer, S. et al. Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science 362, 1285–1288 (2018).
Janda, K. D., Schloeder, D., Benkovic, S. J. & Lerner, R. A. Induction of an antibody that catalyzes the hydrolysis of an amide bond. Science 241, 1188–1191 (1988).
Thayer, M. M. et al. Structural basis for amide hydrolysis catalyzed by the 43C9 antibody. J. Mol. Biol. 291, 329–345 (1993).
Roberts, V. A., Stewart, J., Benkovic, S. J. & Getzoff, E. D. Catalytic antibody model and mutagenesis implicate arginine in transition-state stabilization. J. Mol. Biol. 235, 1098–1116 (1994).
Doyle, A. G. & Jacobsen, E. N. Small-molecule H-bond donors in asymmetric catalysis. Chem. Rev. 107, 5713–5743 (2007).
Taylor, M. S. & Jacobsen, E. N. Asymmetric catalysis by chiral hydrogen-bond donors. Angew. Chem. Int. Ed. 45, 1520–1543 (2006).
Amarante, G. W. et al. Brønsted acid catalyzed Morita-Baylis-Hillman reaction: a new mechanistic view for thioureas revealed by ESI‐MS(/MS) monitoring and DFT calculations. Chem. Eur. J. 15, 12460–12469 (2009).
Knowles, R. R., Lin, S. & Jacobsen, E. N. Enantioselective thiourea-catalyzed cationic polycyclizations. J. Am. Chem. Soc. 132, 5030–5032 (2010).
Park, Y. et al. Macrocyclic bis-thioureas catalyze stereospecific glycosylation reactions. Science 355, 162–166 (2017).
St-Jacques, A. D., Eyahpaise, M.-E. C. & Chica, R. A. Computational design of multisubstrate enzyme specificity. ACS Catal. 9, 5480–5485 (2019).
Davey, J. A., Damry, A. M., Goto, N. K. & Chica, R. A. Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol. 13, 1280–1285 (2017).
Lee, T. S. et al. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 5, 12 (2011).
Kille, S. et al. Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth. Biol. 2, 83–92 (2013).
Luo, S., Wang, P. G. & Cheng, J.-P. Remarkable rate acceleration of imidazole-promoted Baylis-Hillman reaction involving cyclic enones in basic water solution. J. Org. Chem. 69, 555–558 (2004).
Kataoka, T., Iwama, T., Tsujiyama, S., Iwamura, T. & Watanabe, S. The Chalcogeno-Baylis-Hillman reaction: a new preparation of allylic alcohols from aldehydes and electron-deficient alkenes. Tetrahedron 54, 11813–11824 (1998).
Vazquez-Chavez, J. et al. Effect of chiral N-substituents with methyl and trifluoromethyl groups on the catalytic performance of mono- and bifunctional thioureas. Org. Biomol. Chem. 17, 10045–10051 (2019).
Wang, F. et al. A highly efficient kinetic resolution of Morita-Baylis-Hillman adducts achieved by N-Ar axially chiral Pd-complexes catalyzed asymmetric allylation. Chem. Commun. 47, 12813–12815 (2011).
Kwong, C. K.-W., Huang, R., Zhang, M., Shi, M. & Toy, P. H. Bifunctional polymeric organocatalysts and their application in the cooperative catalysis of Morita-Baylis-Hillman reaction. Chemistry 13, 2369–2376 (2007).
Li, G., Wei, H.-X., Gao, J. J. & Caputo, T. D. TiCl4-mediated Baylis-Hillman and aldol reactions without the direct use of a Lewis base. Tetrahedron Lett. 41, 1–5 (2000).
Yang, J. et al. Endohedral functionalized cage as a tool to create frustrated Lewis pairs. Angew. Chem. Int. Ed. 57, 14212–14215 (2018).
Venable, J. D. et al. Preparation and biological evaluation of indole, benzimidazole, and thienopyrrole piperazine carboxamides: potent human histamine H4 antagonists. J. Med. Chem. 48, 8289–8298 (2005).
Comins, D. L. & Killpack, M. O. Lithiation of heterocycles directed by α-amino alkoxides. J. Org. Chem. 52, 104–109 (1987).
Coelho, F. et al. Ultrasound in Baylis-Hillman reactions with aliphatic and aromatic aldehydes: scope and limitations. Tetrahedron 58, 7437–7447 (2002).
Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D 74, 85–97 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Frisch, M. J. et al. Gaussian 16, Revision A.03 (Gaussian, 2016).
Becke, A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Hehre, W., Ditchfield, R. & Pople, J. Further extensions of Gaussian-type basis sets for use in molecular orbital studies of organic molecules. J. Chem. Phys. 56, 2257–2261 (1972).
Francl, M. et al. Self-consistent molecular orbital methods. XXIII. A polarization-type basis set for 2nd-row elements. J. Chem. Phys. 77, 3654–3665 (1982).
Grimme, S., Ehrlich, S. & Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 32, 1456–1465 (2011).
Barone, V. & Cossi, M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J. Phys. Chem. 102, 1995–2001 (1998).
Cossi, M., Rega, N., Scalmani, G. & Barone, V. Energies, structures, and electronic properties of molecules in solution with the C‐PCM solvation model. J. Comput. Chem. 24, 669–681 (2003).
Shaik, S., Kumar, D., de Visser, S. P., Altun, A. & Thiel, W. Theoretical perspective on the structure and mechanism of cytochrome P450 enzymes. Chem. Rev. 105, 2279–2328 (2005).
Heyes, D. J., Sakuma, M., de Visser, S. P. & Scrutton, N. S. Nuclear quantum tunneling in the light-activated enzyme protochlorophyllide oxidoreductase. J. Biol. Chem. 284, 3762–3767 (2009).
Acknowledgements
We acknowledge the Biotechnology and Biological Sciences Research Council (David Phillips Fellowship BB/M027023/1 to A.P.G.), the European Research Council (ERC Starter Grant no. 757991 to A.P.G.) and the UK Research and Innovation Council (Future Leader Fellowship MR/T041722/1 to S.L.L.). We thank the Faculty of Science and Engineering (University of Manchester) for the award of a Presidential Fellowship to S.L.L. A.E.C. was supported by a BBSRC Industrial CASE PhD studentship (BB/S507040/1) supported by GSK. We are grateful to the Diamond Light Source for time on beamlines i03 and i04 under proposals MX17773–33 and MX17773–74, to the Manchester SYNBIOCHEM Centre (BB/M017702/1), the Future Biomanufacturing Hub (EP/S01778X/1) and the Henry Royce Institute for Advanced Materials (funded through EPSRC grants nos. EP/R00661X/1, EP/S019367/1, EP/P025021/1 and EP/P025498/1) for access to their facilities, and to M. Dunstan (Manchester Institute of Biotechnology) for guidance on automating directed evolution workflows. We thank R. Spiess and R. Sung (Manchester Institute of Biotechnology) for acquiring protein mass spectra and for assistance with HPLC method development, and Reach Separations (Nottingham) for supplying individual enantiomers of MBH adduct 3. We acknowledge assistance given by IT Services and use of the Computational Shared Facility at the University of Manchester.
Author information
Authors and Affiliations
Contributions
R.C. carried out molecular biology, protein production, purification, crystallization and kinetic characterization, and directed evolution experiments. A.E.C. and R.C. carried out organic synthesis and substrate profiling of the BH32 variants. R.C., A.J.B. and A.E.C. developed spectrophotometric assays and performed enzyme-inhibition experiments. L.J. and S.H. carried out molecular-docking and DFT calculations. S.H. interpreted and analysed kinetic data. C.L. interpreted, analysed and presented structural data. D.B. provided the BH32 design model. All authors discussed the results and participated in writing the manuscript. A.P.G. and S.L.L. directed the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemistry thanks Andrew Buller, Elaine O’Reilly and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13, Tables 1–9, DNA and protein sequences, Cartesian coordinates for energy-minimized cluster models.
Supplementary Data 1
Source data for Supplementary Figs. 2, 5, 6, 11 and 13.
Supplementary Data 2
Raw NMR data for Supplementary Fig. 4a.
Supplementary Data 3
Raw NMR data for Supplementary Fig. 4b.
Source data
Source Data Fig. 1
Raw data supporting the main text and Fig. 1b.
Source Data Fig. 2
Raw data supporting the main text and Figs. 2b, 2c and 2d.
Rights and permissions
About this article
Cite this article
Crawshaw, R., Crossley, A.E., Johannissen, L. et al. Engineering an efficient and enantioselective enzyme for the Morita–Baylis–Hillman reaction. Nat. Chem. 14, 313–320 (2022). https://doi.org/10.1038/s41557-021-00833-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00833-9
This article is cited by
-
A non-canonical nucleophile unlocks a new mechanistic pathway in a designed enzyme
Nature Communications (2024)
-
The road to fully programmable protein catalysis
Nature (2022)
-
Buy one, get one free
Nature Synthesis (2022)