Abstract
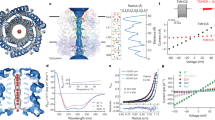
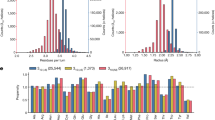
The design of peptides that assemble in membranes to form functional ion channels is challenging. Specifically, hydrophobic interactions must be designed between the peptides and at the peptide–lipid interfaces simultaneously. Here, we take a multi-step approach towards this problem. First, we use rational de novo design to generate water-soluble α-helical barrels with polar interiors, and confirm their structures using high-resolution X-ray crystallography. These α-helical barrels have water-filled lumens like those of transmembrane channels. Next, we modify the sequences to facilitate their insertion into lipid bilayers. Single-channel electrical recordings and fluorescent imaging of the peptides in membranes show monodisperse, cation-selective channels of unitary conductance. Surprisingly, however, an X-ray structure solved from the lipidic cubic phase for one peptide reveals an alternative state with tightly packed helices and a constricted channel. To reconcile these observations, we perform computational analyses to compare the properties of possible different states of the peptide.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
X-ray crystal structures have been submitted to the RCSB Protein Data Bank with accession codes 6YAZ, 6YB0, 6YB1 and 6YB2. Data supporting the results and conclusions are available within this paper and the Supplementary Information. Additional raw data are available at Figshare, https://doi.org/10.6084/m9.figshare.14406419.
References
Huang, P. S., Boyken, S. E. & Baker, D. The coming of age of de novo protein design. Nature 537, 320–327 (2016).
Korendovych, I. V. & DeGrado, W. F. De novo protein design, a retrospective. Q. Rev. Biophys. 53, e3 (2020).
Lu, P. L. et al. Accurate computational design of multipass transmembrane proteins. Science 359, 1042–1046 (2018).
Mravic, M. et al. Packing of apolar side chains enables accurate design of highly stable membrane proteins. Science 363, 1418–1423 (2019).
Joh, N. H. et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524 (2014).
Davey, J. A., Damry, A. M., Goto, N. K. & Chica, R. A. Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol. 13, 1280–1285 (2017).
Chen, K.-Y. M., Keri, D. & Barth, P. Computational design of G Protein-Coupled Receptor allosteric signal transductions. Nat. Chem. Biol. 16, 77–86 (2020).
Woolfson, D. N. Coiled-coil design: updated and upgraded. Subcell. Biochem. 82, 35–61 (2017).
Lear, J. D., Wasserman, Z. R. & Degrado, W. F. Synthetic amphiphilic peptide models for protein ion channels. Science 240, 1177–1181 (1988).
Mahendran, K. R. et al. A monodisperse transmembrane α-helical peptide barrel. Nat. Chem. 9, 411–419 (2017).
Bowie, J. U. Helix packing in membrane proteins. J. Mol. Biol. 272, 780–789 (1997).
Hong, H. Toward understanding driving forces in membrane protein folding. Arch. Biochem. Biophys. 564, 297–313 (2014).
Liu, J. et al. A seven-helix coiled coil. Proc. Natl Acad. Sci. USA 103, 15457–15462 (2006).
Zaccai, N. R. et al. A de novo peptide hexamer with a mutable channel. Nat. Chem. Biol. 7, 935–941 (2011).
Thomson, A. R. et al. Computational design of water-soluble α-helical barrels. Science 346, 485–488 (2014).
Rhys, G. G. et al. Maintaining and breaking symmetry in homomeric coiled-coil assemblies. Nat. Commun. 9, 4132 (2018).
Wood, C. W. & Woolfson, D. N. CCBuilder 2.0: powerful and accessible coiled-coil modeling. Protein Sci. 27, 103–111 (2018).
Walshaw, J. & Woolfson, D. N. SOCKET: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 307, 1427–1450 (2001).
Klesse, G., Rao, S. L., Sansom, M. S. P. & Tucker, S. J. CHAP: a versatile tool for the structural and functional annotation of ion channel pores. J. Mol. Biol. 431, 3353–3365 (2019).
Aryal, P., Sansom, M. S. P. & Tucker, S. J. Hydrophobic gating in ion channels. J. Mol. Biol. 427, 121–130 (2015).
Carugo, O. Statistical survey of the buried waters in the Protein Data Bank. Amino Acids 48, 193–202 (2016).
Dawson, J. P., Weinger, J. S. & Engelman, D. M. Motifs of serine and threonine can drive association of transmembrane helices. J. Mol. Biol. 316, 799–805 (2002).
Hessa, T. et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 (2007).
Harriss, L. M., Cronin, B., Thompson, J. R. & Wallace, M. I. Imaging multiple conductance states in an alamethicin pore. J. Am. Chem. Soc. 133, 14507–14509 (2011).
Landau, E. M. & Rosenbusch, J. P. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 93, 14532–14535 (1996).
Gernert, K. M., Surles, M. C., Labean, T. H., Richardson, J. S. & Richardson, D. C. The Alacoil: a very tight, antiparallel coiled-coil of helices. Protein Sci. 4, 2252–2260 (1995).
Adamian, L. & Liang, J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins 47, 209–218 (2002).
Zhang, S. Q. et al. The membrane- and soluble-protein helix-helix Interactome: similar geometry via different interactions. Structure 23, 527–541 (2015).
Rhys, G. G. et al. Navigating the structural landscape of de novo α-helical bundles. J. Am. Chem. Soc. 141, 8787–8797 (2019).
Song, C. et al. Crystal structure and functional mechanism of a human antimicrobial membrane channel. Proc. Natl Acad. Sci. USA 110, 4586–4591 (2013).
Hayouka, Z. et al. Quasiracemate crystal structures of magainin 2 derivatives support the functional significance of the phenylalanine zipper motif. J. Am. Chem. Soc. 137, 11884–11887 (2015).
Kurgan, K. W. et al. Retention of native quaternary structure in racemic melittin crystals. J. Am. Chem. Soc. 141, 7704–7708 (2019).
Sansom, M. S. The biophysics of peptide models of ion channels. Prog. Biophys. Mol. Biol. 55, 139–235 (1991).
Hille, B. Ionic Channels of Excitable Membranes (Oxford Univ. Press, 2001).
Kienker, P. K., DeGrado, W. F. & Lear, J. D. A helical-dipole model describes the single-channel current rectification of an uncharged peptide ion channel. Proc. Natl Acad. Sci. USA 91, 4859–4863 (1994).
Noskov, S. Y., Im, W. & Roux, B. Ion permeation through the α-hemolysin channel: theoretical studies based on Brownian dynamics and Poisson-Nernst-Plank electrodiffusion theory. Biophys. J. 87, 2299–2309 (2004).
Wang, S. Q., Song, L. S., Lakatta, E. G. & Cheng, H. P. Ca2+ signalling between single L-type Ca2+ channels and ryanodine receptors in heart cells. Nature 410, 592–596 (2001).
Heron, A. J., Thompson, J. R., Cronin, B., Bayley, H. & Wallace, M. I. Simultaneous measurement of ionic current and fluorescence from single protein pores. J. Am. Chem. Soc. 131, 1652–1653 (2009).
Leptihn, S. et al. Constructing droplet interface bilayers from the contact of aqueous droplets in oil. Nat. Protoc. 8, 1048–1057 (2013).
Ramadurai, S., Duurkens, R., Krasnikov, V. V. & Poolman, B. Lateral diffusion of membrane proteins: consequences of hydrophobic mismatch and lipid composition. Biophys. J. 99, 1482–1489 (2010).
Saffman, P. G. & Delbrück, M. Brownian motion in biological membranes. Proc. Natl Acad. Sci. USA 72, 3111–3113 (1975).
Callenberg, K. M. et al. APBSmem: a graphical interface for electrostatic calculations at the membrane. PLoS One 5, e12722 (2010).
Roux, B., Allen, T., Berneche, S. & Im, W. Theoretical and computational models of biological ion channels. Q. Rev. Biophys. 37, 15–103 (2004).
Krishnan, R. S. et al. Autonomously assembled synthetic transmembrane peptide pore. J. Am. Chem. Soc. 141, 2949–2959 (2019).
Spruijt, E., Tusk, S. E. & Bayley, H. DNA scaffolds support stable and uniform peptide nanopores. Nat. Nanotechnol. 13, 739–745 (2018).
Grigoryan, G., Reinke, A. W. & Keating, A. E. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature 458, 859–864 (2009).
Peraro, M. D. & van der Goot, F. G. Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 (2016).
Niitsu, A., Heal, J. W., Fauland, K., Thomson, A. R. & Woolfson, D. N. Membrane-spanning α-helical barrels as tractable protein-design targets. Philos. Trans. Royal Soc. B 372, 20160213 (2017).
Xu, C. et al. Computational design of transmembrane pores. Nature 585, 129–134 (2020).
Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 12, 619–630 (2017).
Dou, J. Y. et al. De novo design of a fluorescence-activating β-barrel. Nature 561, 485–491 (2018).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D 67, 282–292 (2011).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Sammito, M. et al. ARCIMBOLDO_LITE: single-workstation implementation and use. Acta Crystallogr. D 71, 1921–1930 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Maglia, G., Heron, A. J., Stoddart, D., Japrung, D. & Bayley, H. Analysis of single nucleic acid molecules with protein nanopores. Methods Enzymol. 475, 591–623 (2010).
Montal, M. & Mueller, P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl Acad. Sci. USA 69, 3561–3566 (1972).
Gu, L. Q. et al. Reversal of charge selectivity in transmembrane protein pores by using noncovalent molecular adapters. Proc. Natl Acad. Sci. USA 97, 3959–3964 (2000).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Tinevez, J. Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Ramadurai, S. et al. Lateral diffusion of membrane proteins. J. Am. Chem. Soc. 131, 12650–12656 (2009).
Kučerka, N., Nieh, M.-P. & Katsaras, J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta Biomembr. 1808, 2761–2771 (2011).
Acknowledgements
A.J.S. thanks Diamond Light Source for a place on the CCP4 Data Collection and Structure Solution Workshop 2017. E.J.M.L. thanks S. Rao and G. Klesse (University of Oxford) for support with CHAP and F. Marcoline (UCSF) for support with APBSmem. A.J.S. was funded by the Bristol Chemical Synthesis Centre for Doctoral Training funded by the EPSRC (EP/G036764/1). A.N. and K.R.M. were supported by a BBSRC grant to R.L.B., H.B. and D.N.W. (BB/J009784/1). W.M.D., A.N., A.J.S., A.R.T. and D.N.W. were funded by ERC Grants to D.N.W. (340764 and 787173). E.J.M.L. was in the BBSRC/EPSRC-funded Synthetic Biology Research Centre, BrisSynBio (BB/L01386X/1). M.I.W. was funded by the BBSRC (BB/R001790/1). W.F.D. was supported by NIH (R35 GM122603), NSF (1709506) and US Air Force (1709506) grants. H.T.K. was supported by the NIH Ruth L. Kirschstein NRSA Postdoctoral Fellowship (F32 GM125217). M.M. is supported by the Howard Hughes Medical Institute Gilliam Fellowship. D.N.W. held a Royal Society Wolfson Research Merit Award (WM140008). Beamline 8.3.1 at the Advanced Light Source is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives grant MR-15-328599, the National Institutes of Health (R01 GM124149 and P30 GM124169), Plexxikon Inc. and the Integrated Diffraction Analysis Technologies program of the US Department of Energy Office of Biological and Environmental Research. The Advanced Light Source (Berkeley, CA) is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under contract number DE-AC02-05CH11231, Office of Basic Energy Sciences.
Author information
Authors and Affiliations
Contributions
A.J.S., A.N., K.R.M., A.R.T., H.B., R.L.B. and D.N.W. conceived the project and designed the peptides, which A.J.S., A.N. and W.M.D. synthesized. A.J.S., A.N. and K.R.M performed solution-phase biophysics and electrophysiology experiments. A.J.S., H.T.K. and L.L. determined X-ray crystal structures. A.J.S. and A.R.T. did the computational design. E.J.M.L., M.M. and A.J.M. ran and analysed the MD simulations. E.J.M.L. conducted the electrostatic calculations. J.T.S. and M.I.W. conducted and analysed the oSCR. A.J.S. and A.N. contributed equally as first authors to this work. The contributions of H.T.K., E.J.M.L. and J.T.S. were also equal. A.J.S., E.J.M.L. and D.N.W. wrote the manuscript, to which all authors contributed.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Computational methods, Supplementary Figs. 1–67, Tables 1–5, Video captions 1–5 and refs. 1–36.
Supplementary Video 1
Molecular dynamics simulation of CC-Type2-TI5.
Supplementary Video 2
Molecular dynamics simulation of CC-Type2-LIS4.
Supplementary Video 3
Molecular dynamics simulation of CC-Type2-TS2.
Supplementary Video 4
oSCR recording of CCTM-VI channels..
Supplementary Video 5
Molecular dynamics simulation of K2-CCTM-VI.
Rights and permissions
About this article
Cite this article
Scott, A.J., Niitsu, A., Kratochvil, H.T. et al. Constructing ion channels from water-soluble α-helical barrels. Nat. Chem. 13, 643–650 (2021). https://doi.org/10.1038/s41557-021-00688-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00688-0
This article is cited by
-
Sparks of function by de novo protein design
Nature Biotechnology (2024)
-
Microbial membrane transport proteins and their biotechnological applications
World Journal of Microbiology and Biotechnology (2024)
-
Transient water wires mediate selective proton transport in designed channel proteins
Nature Chemistry (2023)
-
Differential sensing with arrays of de novo designed peptide assemblies
Nature Communications (2023)
-
Segmentation strategy of de novo designed four-helical bundles expands protein oligomerization modalities for cell regulation
Nature Communications (2023)