Abstract
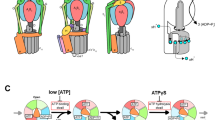
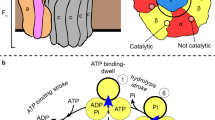
The ATPase-catalysed conversion of ATP to ADP is a fundamental process in biology. During the hydrolysis of ATP, the α3β3 domain undergoes conformational changes while the central stalk (γ/D) rotates unidirectionally. Experimental studies have suggested that different catalytic mechanisms operate depending on the type of ATPase, but the structural and energetic basis of these mechanisms remains unclear. In particular, it is not clear how the positions of the catalytic dwells influence the energy transduction. Here we show that the observed dwell positions, unidirectional rotation and movement against the applied torque are reflections of the free-energy surface of the systems. Instructively, we determine that the dwell positions do not substantially affect the stopping torque. Our results suggest that the three resting states and the pathways that connect them should not be treated equally. The current work demonstrates how the free-energy landscape determines the behaviour of different types of ATPases.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The cryo-EM structures used in this work are available from the Protein Data Bank under PDB IDs 6RD9, 6RDC and 6REP for mF1; 6FKF, 6FKI and 6FKH for cF1; and 6QUM, 6R0W and 6R0Y for V1. The CG structure and input codes for CG relaxation and MCPT calculation are available freely online: https://doi.org/10.6084/m9.figshare.12768413.v2
Code availability
The MOLARIS-XG package is available upon request from the University of Southern California.
References
Boyer, P. D. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 (1997).
Kühlbrandt, W. Structure and mechanisms of F-type ATP synthases. Annu. Rev. Biochem. 88, 515–549 (2019).
Walker, J. E. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16 (2013).
Walker, J. E. ATP synthesis by rotary catalysis (Nobel lecture). Angew. Chem. Int. Ed. 37, 2308–2319 (1998).
Junge, W. & Nelson, N. ATP synthase. Annu. Rev. Biochem. 84, 631–657 (2015).
Mukherjee, S. & Warshel, A. The FOF1 ATP synthase: from atomistic three-dimensional structure to the rotary-chemical function. Photosynth. Res. 134, 1–15 (2017).
Watanabe, R. et al. Mechanical modulation of catalytic power on F1-ATPase. Nat. Chem. Biol. 8, 86–92 (2012).
Adachi, K. et al. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 (2007).
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K. & Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001).
Masaike, T., Koyama-Horibe, F., Oiwa, K., Yoshida, M. & Nishizaka, T. Cooperative three-step motions in catalytic subunits of F1-ATPase correlate with 80° and 40° substep rotations. Nat. Struct. Mol. Biol. 15, 1326–1333 (2008).
Suzuki, T., Tanaka, K., Wakabayashi, C., Saita, E. & Yoshida, M. Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat. Chem. Biol. 10, 930–936 (2014).
Bason, J. V., Montgomery, M. G., Leslie, A. G. W. & Walker, J. E. How release of phosphate from mammalian F1-ATPase generates a rotary substep. Proc. Natl Acad. Sci. USA 112, 6009–6014 (2015).
Kobayashi, R., Ueno, H., Li, C.-B. & Noji, H. Rotary catalysis of bovine mitochondrial F1-ATPase studied by single-molecule experiments. Proc. Natl Acad. Sci. USA 117, 1447–1456 (2020).
Suzuki, K. et al. Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor. Nat. Commun. 7, 13235 (2016).
Minagawa, Y. et al. Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1-ATPase. J. Biol. Chem. 288, 32700–32707 (2013).
Murphy, B. J. et al. Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1–Fo coupling. Science 364, eaaw9128 (2019).
Hahn, A., Vonck, J., Mills, D. J., Meier, T. & Kühlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360, 4318 (2018).
Zhou, L. & Sazanov, L. A. Structure and conformational plasticity of the intact Thermus thermophilus V/A-type ATPase. Science 365, eaaw9144 (2019).
Majewski, D. D. et al. Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry. Nat. Commun. 10, 626 (2019).
Srivastava, A. P. et al. High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Science 360, eaas9699 (2018).
Sielaff, H., Yanagisawa, S., Frasch, W. D., Junge, W. & Börsch, M. Structural asymmetry and kinetic limping of single rotary F-ATP synthases. Molecules 24, 504 (2019).
Mukherjee, S. & Warshel, A. Electrostatic origin of the mechanochemical rotary mechanism and the catalytic dwell of F1-ATPase. Proc. Natl Acad. Sci. USA 108, 20550–20555 (2011).
Dittrich, M., Hayashi, S. & Schulten, K. On the mechanism of ATP hydrolysis in F1-ATPase. Biophys. J. 85, 2253–2266 (2003).
S̆trajbl, M., Shurki, A. & Warshel, A. Converting conformational changes to electrostatic energy in molecular motors: the energetics of ATP synthase. Proc. Natl Acad. Sci. USA 100, 14834–14839 (2003).
Mukherjee, S. & Warshel, A. Realistic simulations of the coupling between the protomotive force and the mechanical rotation of the F0-ATPase. Proc. Natl Acad. Sci. USA 109, 14876–14881 (2012).
Lee, M., Kolev, V. & Warshel, A. Validating a coarse-grained voltage activation model by comparing its performance to the results of monte carlo simulations. J. Phys. Chem. B 121, 11284–11291 (2017).
Vorobyov, I., Kim, I., Chu, Z. T. & Warshel, A. Refining the treatment of membrane proteins by coarse-grained models. Proteins Struct. Funct. Bioinformatics 84, 92–117 (2016).
Vicatos, S., Rychkova, A., Mukherjee, S. & Warshel, A. An effective coarse-grained model for biological simulations: recent refinements and validations. Proteins 82, 1168–1185 (2014).
Zhang, Z. & Thirumalai, D. Dissecting the kinematics of the kinesin step. Structure 20, 628–640 (2012).
Alhadeff, R. & Warshel, A. Reexamining the origin of the directionality of myosin V. Proc. Natl Acad. Sci. USA 114, 10426–10431 (2017).
Warshel, A., Sharma, P. K., Kato, M. & Parson, W. W. Modeling electrostatic effects in proteins. Biochim. Biophys. Acta Proteins Proteom. 1764, 1647–1676 (2006).
Bai, C. & Warshel, A. Revisiting the protomotive vectorial motion of F0-ATPase. Proc. Natl Acad. Sci. USA, 116, 19484–19489 (2019).
Alhadeff, R. & Warshel, A. A free-energy landscape for the glucagon-like peptide 1 receptor GLP1R. Proteins Struct. Funct. Bioinformatics 88, 127–134 (2020).
Kim, I. & Warshel, A. Analyzing the electrogenicity of cytochrome c oxidase. Proc. Natl Acad. Sci. USA 113, 7810–7815 (2016).
Mukherjee, S. & Warshel, A. Dissecting the role of the γ-subunit in the rotary–chemical coupling and torque generation of F1-ATPase. Proc. Natl Acad. Sci. USA 112, 2746–2751 (2015).
Mukherjee, S., Bora, R. P. & Warshel, A. Torque, chemistry and efficiency in molecular motors: a study of the rotary–chemical coupling in F1-ATPase. Q. Rev. Biophys. 48, 395–403 (2015).
Astumian, R. D., Mukherjee, S. & Warshel, A. The physics and physical chemistry of molecular machines. ChemPhysChem 17, 1719–1741 (2016).
Ueno, H. et al. Torque generation of Enterococcus hirae V-ATPase. J. Biol. Chem. 289, 31212–31223 (2014).
Pänke, O., Cherepanov, D. A., Gumbiowski, K., Engelbrecht, S. & Junge, W. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 81, 1220–1233 (2001).
Arai, HidenobuC. et al. Torque generation mechanism of F1-ATPase upon NTP binding. Biophys. J. 107, 156–164 (2014).
Sakaki, N. et al. One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 88, 2047–2056 (2005).
McMillan, D. G. G., Watanabe, R., Ueno, H., Cook, G. M. & Noji, H. Biophysical characterization of a thermoalkaliphilic molecular motor with a high stepping torque gives insight into evolutionary ATP synthase adaptation. J. Biol. Chem. 291, 23965–23977 (2016).
Hisabori, T., Kondoh, A. & Yoshida, M. The γ subunit in chloroplast F1-ATPase can rotate in a unidirectional and counter-clockwise manner. FEBS Lett. 463, 35–38 (1999).
Yasuda, R., Noji, H., Kinosita, K. & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell 93, 1117–1124 (1998).
Noji, H., Ueno, H. & McMillan, D. G. G. Catalytic robustness and torque generation of the F1-ATPase. Biophys. Rev. 9, 103–118 (2017).
Imamura, H. et al. Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl Acad. Sci. USA 102, 17929–17933 (2005).
Astumian, R. D. Kinetic asymmetry allows macromolecular catalysts to drive an information ratchet. Nat. Commun. 10, 3837 (2019).
Astumian, R. D. Thermodynamics and kinetics of molecular motors. Biophys. J. 98, 2401–2409 (2010).
Gao, Y. Q., Yang, W. & Karplus, M. A structure-based model for the synthesis and hydrolysis of ATP by F1-ATPase. Cell 123, 195–205 (2005).
Kamerlin, S. C. L., Vicatos, S., Dryga, A. & Warshel, A. Coarse-grained (multiscale) simulations in studies of biophysical and chemical systems. Annu. Rev. Phys. Chem. 62, 41–64 (2011).
Lee, F. S., Chu, Z. T. & Warshel, A. Microscopic and semimicroscopic calculations of electrostatic energies in proteins by the POLARIS and ENZYMIX programs. J. Comput. Chem. 14, 161–185 (1993).
Robert, C. P. & Casella, G. Monte Carlo Statistical Methods (Springer, 2005).
Torrie, G. M. & Valleau, J. P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 23, 187–199 (1977).
Acknowledgements
This work was supported by the National Science Foundation grant MCB 1707167 and the National Institute of Health grant R01-AI055926. We thank the University of Southern California High Performance Computing and Communication Center (HPCC), as well as the Extreme Science and Engineering Discovery Environment’s (XSEDE) Comet facility at the San Diego Supercomputing Center, for computational resources.
Author information
Authors and Affiliations
Contributions
C.B. and A.W. conceived and designed the project. C.B. designed, executed and analysed all experiments with guidance from A.W. M.A. executed and analysed part of the experiments with assistance and guidance from C.B. and A.W. C.B. wrote the input and analysis codes for all experiments. C.B. and A.W. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary Figs. 1 and 2 and discussion.
Rights and permissions
About this article
Cite this article
Bai, C., Asadi, M. & Warshel, A. The catalytic dwell in ATPases is not crucial for movement against applied torque. Nat. Chem. 12, 1187–1192 (2020). https://doi.org/10.1038/s41557-020-0549-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-020-0549-6