Abstract
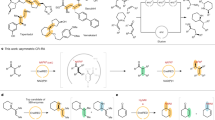
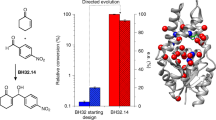
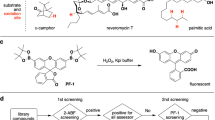
Finding faster and simpler ways to screen protein sequence space to enable the identification of new biocatalysts for asymmetric synthesis remains both a challenge and a rate-limiting step in enzyme discovery. Biocatalytic strategies for the synthesis of chiral amines are increasingly attractive and include enzymatic asymmetric reductive amination, which offers an efficient route to many of these high-value compounds. Here we report the discovery of over 300 new imine reductases and the production of a large (384 enzymes) and sequence-diverse panel of imine reductases available for screening. We also report the development of a facile high-throughput screen to interrogate their activity. Through this approach we identified imine reductase biocatalysts capable of accepting structurally demanding ketones and amines, which include the preparative synthesis of N-substituted β-amino ester derivatives via a dynamic kinetic resolution process, with excellent yields and stereochemical purities.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data supporting the results and conclusions are available within this paper and the Supplementary Information. Both the IREDy-to-go screen and a duplication of the screening plate without the colorimetric screening components containing 0.5 mg of crude lysate of each IRED in each well are freely available through Prozomix Ltd.
References
Kan, S. B. J., Lewis, R. D., Chen, K. & Arnold, F. H. Directed evolution of cytochrome C for carbon–silicon bond formation: bringing silicon to life. Science 354, 1048–1051 (2016).
Wijma, H. J. et al. Enantioselective enzymes by computational design and in silico screening. Angew. Chem. Int. Ed. 54, 3726–3730 (2015).
Burke, A. J. et al. Design and evolution of an enzyme with a non-canonical organocatalytic mechanism. Nature 570, 219–223 (2019).
Zeymer, C., Zschoche, R. & Hilvert, D. Optimization of enzyme mechanism along the evolutionary trajectory of a computationally designed (retro-)aldolase. J. Am. Chem. Soc. 139, 12541–12549 (2017).
Sandoval, B. A., Meichan, A. J. & Hyster, T. K. Enantioselective hydrogen atom transfer: discovery of catalytic promiscuity in flavin-dependent ‘ene’-reductases. J. Am. Chem. Soc. 139, 11313–11316 (2017).
Sandoval, B. A. & Hyster, T. K. Emerging strategies for expanding the toolbox of enzymes in biocatalysis. Curr. Opin. Chem. Biol. 55, 45–51 (2020).
Devine, P. N. et al. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2, 409–421 (2018).
Xiao, H., Bao, Z. & Zhao, H. High throughput screening and selection methods for directed enzyme evolution. Ind. Eng. Chem. Res. 54, 4011–4020 (2015).
Robbins, D. W. & Hartwig, J. F. A simple, multidimensional approach to high-throughput discovery of catalytic reactions. Science 333, 1423–1427 (2011).
Friedfeld, M. R. et al. Cobalt precursors for high-throughput discovery of base metal asymmetric alkene hydrogenation catalysts. Science 342, 1076 (2013).
Collins, K. D., Gensch, T. & Glorius, F. Contemporary screening approaches to reaction discovery and development. Nat. Chem. 6, 859–871 (2014).
Cabrera-Pardo, J., Chai, D. I., Liu, S., Mrksich, M. & A. Kozmin, S. Label-assisted mass spectrometry for the acceleration of reaction discovery and optimization. Nat. Chem. 5, 423–427 (2013).
Vanacek, P. et al. Exploration of enzyme diversity by integrating bioinformatics with expression analysis and biochemical characterization. ACS Catal. 8, 2402–2412 (2018).
Yan, C. et al. Real-time screening of biocatalysts in live bacterial colonies. J. Am. Chem. Soc. 139, 1408–1411 (2017).
Vallejo, D., Nikoomanzar, A., Paegel, B. M. & Chaput, J. C. Fluorescence-activated droplet sorting for single-cell directed evolution. ACS Synth. Biol. 8, 1430–1440 (2019).
Gielen, F. et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proc. Natl Acad. Sci. USA 113, E7383–E7389 (2016).
Davids, T., Schmidt, M., Böttcher, D. & Bornscheuer, U. T. Strategies for the discovery and engineering of enzymes for biocatalysis. Curr. Opin. Chem. Biol. 17, 215–220 (2013).
Bunzel, H. A., Garrabou, X., Pott, M. & Hilvert, D. Speeding up enzyme discovery and engineering with ultrahigh-throughput methods. Curr. Opin. Struct. Biol. 48, 149–156 (2018).
Ghislieri, D. & Turner, N. J. Biocatalytic approaches to the synthesis of enantiomerically pure chiral amines. Top. Catal. 57, 284–300 (2014).
Weise, N., Parmeggiani, F., Ahmed, S. & Turner, N. J. Discovery and investigation of mutase-like activity in a phenylalanine ammonia lyase from Anabaena variabilis. Top. Catal. 61, 288–295 (2018).
Grogan, G. Synthesis of chiral amines using redox biocatalysis. Curr. Opin. Chem. Biol. 43, 15–22 (2018).
Turner, N. J. Enantioselective oxidation of C–O and C–N bonds using oxidases. Chem. Rev. 111, 4073–4087 (2011).
Jia, Z.-J., Gao, S. & Arnold, F. H. Enzymatic primary amination of benzylic and allylic C(sp3)-H bonds. J. Am. Chem. Soc. 142, 10279–10283 (2020).
Schrittwieser, J. H., Velikogne, S. & Kroutil, W. Biocatalytic imine reduction and reductive amination of ketones. Adv. Synth. Catal. 357, 1665–1685 (2015).
Hyslop, J. F. et al. Biocatalytic synthesis of chiral N-functionalized amino acids. Angew. Chem. Int. Ed. 57, 13821–13824 (2018).
Stahl, L. The Handbook of Homogeneous Hydrogenation, Volumes 1−3 Edited by Johannes G. de Vries (DSM Pharmaceutical Products, Geleen, The Netherlands) and Cornelis J. Elsevier (Universiteit van Amsterdam, The Netherlands). Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim. 2007. 1632 pp. $625.00. ISBN 978-3-527-31161-3. J. Am. Chem. Soc. 129, 10297–10298 (2007).
Li, W. Stereoselective Formation of Amines (Springer, 2014).
Roughley, S. D. & Jordan, A. M. The medicinal chemist’s toolbox: an analysis of reactions used in the pursuit of drug candidates. J. Med. Chem. 54, 3451–3479 (2011).
Wang, L. & Xiao, J. Hydrogen-atom transfer reactions. Top. Curr. Chem. 374, 1–55 (2016).
Hou, G. et al. Enantioselective hydrogenation of N–H imines. J. Am. Chem. Soc. 131, 9882–9883 (2009).
Aurelio, L., Brownlee, R. & Hughes, A. B. Synthetic preparation of N-methyl-ɑ-amino acids. Chem. Rev. 104, 5823–5846 (2004).
Aleku, A. G. et al. A reductive aminase from Aspergillus oryzae. Nat. Chem. 9, 961–969 (2017).
Tobias, H. et al. Direct reductive amination of ketones: structure and activity of S‐selective imine reductases from Streptomyces. ChemCatChem 6, 2248–2252 (2014).
Scheller, P. N., Lenz, M., Hammer, S. C., Hauer, B. & Nestl, B. M. Imine reductase‐catalyzed intermolecular reductive amination of aldehydes and ketones. ChemCatChem 7, 3239–3242 (2015).
Wetzl, D. et al. Asymmetric reductive amination of ketones catalyzed by imine reductases. ChemCatChem 8, 2023–2026 (2016).
Maugeri, Z. & Rother, D. Reductive amination of ketones catalyzed by whole cell biocatalysts containing imine reductases (IREDs). J. Biotechnol. 258, 167–170 (2017).
Roiban, G. D. et al. Efficient biocatalytic reductive aminations by extending the imine reductase toolbox. ChemCatChem 9, 4475–4479 (2017).
Sharma, M. et al. A mechanism for reductive amination catalyzed by fungal reductive aminases. ACS Catal. 8, 11534–11541 (2018).
Mitsukura, K., Suzuki, M., Tada, K., Yoshida, T. & Nagasawa, T. Asymmetric synthesis of chiral cyclic amine from cyclic imine by bacterial whole-cell catalyst of enantioselective imine reductase. Org. Biomol. Chem. 8, 4533–4535 (2010).
Mitsukura, K. et al. Purification and characterization of a novel (R)-imine reductase from Streptomyces sp. GF3587. Biosci. Biotechnol. Biochem. 75, 1778–1782 (2011).
Slabu, I., Galman, J. L., Lloyd, R. C. & Turner, N. J. Discovery, engineering, and synthetic application of transaminase biocatalysts. ACS Catal. 7, 8263–8284 (2017).
Mangas-Sanchez, J. et al. Imine reductases (IREDs). Curr. Opin. Chem. Biol. 37, 19–25 (2017).
Lenz, M., Borlinghaus, N., Weinmann, L. & Nestl, B. M. Recent advances in imine reductase-catalyzed reactions. World J. Microbiol. Biotechnol. 33, 199 (2017).
Schober, M. et al. Chiral synthesis of LSD1 inhibitor GSK2879552 enabled by directed evolution of an imine reductase. Nat. Catal. 2, 909–915 (2019).
Wetzl, D. et al. Expanding the imine reductase toolbox by exploring the bacterial protein‐sequence space. ChemBioChem 16, 1749–1756 (2015).
France, S. P. et al. Identification of novel bacterial members of the imine reductase enzyme family that perform reductive amination. ChemCatChem 10, 510–514 (2018).
Gheorghe‐Doru, R. et al. Efficient biocatalytic reductive aminations by extending the imine reductase toolbox. ChemCatChem 9, 4475–4479 (2017).
Zhu, J. et al. Enantioselective synthesis of 1-aryl-substituted tetrahydroisoquinolines employing imine reductase. ACS Catal. 7, 7003–7007 (2017).
Li, H., Luan, Z. J., Zheng, G. W. & Xu, J. H. Efficient synthesis of chiral indolines using an imine reductase from Paenibacillus lactis. Adv. Synth. Catal. 357, 1692–1696 (2015).
Nugent, T. C. & El‐Shazly, M. Chiral amine synthesis—recent developments and trends for enamide reduction, reductive amination, and imine reduction. Adv. Synth. Catal. 352, 753–819 (2010).
Baud, D., Jeffries, J. W. E., Moody, T. S., Ward, J. M. & Hailes, H. C. A metagenomics approach for new biocatalyst discovery: application to transaminases and the synthesis of allylic amines. Green Chem. 19, 1134–1143 (2017).
Hernández, K., Szekrenyi, A. & Clapés, P. Nucleophile promiscuity of natural and engineered aldolases. ChemBioChem 19, 1353–1358 (2018).
Leipold, L. et al. The identification and use of robust transaminases from a domestic drain metagenome. Green Chem. 21, 75–86 (2019).
Caparco, A. et al. Metagenomic mining for amine dehydrogenase discovery. Adv. Synth. Catal. 362, 2427 (2020).
Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity Treaty (UK Parliament Foreign And Commonwealth Office, 2010).
Zawodny, W. et al. Chemoenzymatic synthesis of substituted azepanes by sequential biocatalytic reduction and organolithium-mediated rearrangement. J. Am. Chem. Sci. 140, 17872–17877 (2018).
Montgomery, S. L. et al. Characterization of imine reductases in reductive amination for the exploration of structure–activity relationships. Sci. Adv. 6, eaay9320 (2020).
Mayer, K. M. & Arnold, F. H. A colorimetric assay to quantify dehydrogenase activity in crude cell lysates. J. Biomol. Screen 7, 135–140 (2002).
Peng, H. et al. Deciphering piperidine formation in polyketide-derived indolizidines reveals a thioester reduction, transamination, and unusual imine reduction process. ACS Chem. Biol. 11, 3278–3283 (2016).
Ye, S. et al. Identification by genome mining of a type I polyketide gene cluster from Streptomyces argillaceus involved in the biosynthesis of pyridine and piperidine alkaloids argimycins P. Front. Microbiol. 8, 194 (2017).
Lenz, M. et al. Asymmetric ketone reduction by imine reductases. ChemBioChem 18, 253–256 (2017).
González-Martínez, D. et al. Asymmetric synthesis of primary and secondary β-fluoro-arylamines using reductive aminases from fungi. ChemCatChem 12, 2421 (2020).
Smith, J. A. et al. in A101 Advances in Cough, Dyspnea, and Interventional Pulmonary A2672–A2672 (American Thoracic Society, 2017).
Darcsi, A., Tóth, G., Kökösi, J. & Béni, S. Structure elucidation of a process-related impurity of dapoxetine. J. Pharm. Biomed. Anal. 96, 272–277 (2014).
Fuchs, M., Koszelewski, D., Tauber, K., Kroutil, W. & Faber, K. Chemoenzymatic asymmetric total synthesis of (S)-rivastigmine using omega-transaminases. Chemic. Commun. 46, 5500–5502 (2010).
Zhang, D. et al. Development of β-amino acid dehydrogenase for the synthesis of β-amino acids via reductive amination of β-keto acids. ACS Catal. 5, 2220–2224 (2015).
Midelfort, K. et al. Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin. Protein Eng. Des. Sel. 26, 25–33 (2013).
Juraisti, E. & Soloshonok, V. Enantioselective Synthesis of β-Amino Acids 2nd edn (Wiley, 2005).
Iglesias, E. Ester hydrolysis and enol nitrosation reactions of ethyl cyclohexanone-2-carboxylate inhibited by β-cyclodextrin. J. Org. Chem. 65, 6583–6594 (2000).
Bornadel, A. et al. Technical considerations for scale-up of imine-reductase-catalyzed reductive amination: a case study. Org Process. Res. Dev. 23, 1262–1268 (2019).
Mangas-Sanchez, J. et al. Asymmetric synthesis of primary amines catalyzed by fungal reductive aminases. Chem. Sci. 11, 5052–5057 (2020).
Sutin, L. et al. Oxazolones as potent inhibitors of 11β-hydroxysteroid dehydrogenase type 1. Bioorg. Med. Chem. Lett. 17, 4837–4840 (2007).
Afanasyev, O. I., Kuchuk, E., Usanov, D. L. & Chusov, D. Reductive amination in the synthesis of pharmaceuticals. Chem. Rev. 119, 11857–11911 (2019).
Salzmann, T. N., Ratcliffe, R. W., Christensen, B. G. & Bouffard, F. A. A stereocontrolled synthesis of (+)-thienamycin. J. Am. Chem. Sci. 102, 6161–6163 (1980).
Acknowledgements
We thank the Industrial Biotechnology Innovation Centre (IBioIC) and Biotechnology and Biological Sciences Research Council (BBSRC) for awarding the CASE studentship to J.R.M. from Prozomix Ltd. P.Y. was supported by a CSC scholarship and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Grant no. 2016166). T.W.T. was supported by a BBSRC CASE studentship awarded by Pfizer. S.L.M. was supported by a BBSRC CASE studentship from Johnson Matthey. R.B.P. and R.S.H. were supported by the European Research Council (ERC Grant no. 742987). J.M.-S. was funded by grant BB/M006832/1 from the UK Biotechnology and Biological Sciences Research Council. S.J.C., D.J.C., J.D.F., R.A.M.D. and K.M.G. acknowledge the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 685474 for supporting MetaFluidics. N.J.T. is grateful to the ERC for the award of an Advanced Grant (Grant no. 742987). We thank D. Heyes of the Manchester Institute of Biotechnology (MIB) for assistance in gathering the circular dichroism data. We thank Y. Qi of Prozomix Ltd for screening of the Prozomix diaphorases. Prozomix and J.R.M., P.Y., S.L.M., T.W.T., R.B.P., J.M.-S., R.S.H. and N.J.T. also thank other staff of Prozomix for their support.
Author information
Authors and Affiliations
Contributions
N.J.T. and S.J.C. devised and supervised the project. J.M.-S. and R.S.H. managed the project. J.R.M., J.D.F, K.M.G., S.L.M. and D.J.C. performed the identification, cloning and expression of the enzymes. J.R.M., J.D.F., R.A.M.D. and S.J.C. were involved in the design, development and implementation of the colorimetric screen and 384-well plates. J.R.M. and P.Y. carried out the high-throughput characterization of the enzymes. J.R.M. and P.Y. performed the analytical scale biotransformations. P.Y. conducted the preparative-scale biotransformations. P.Y. and R.B.P. synthesized the chemical standards. J.R.M. and T.W.T. undertook the thermostability studies. N.J.T., S.J.C., J.R.M., P.Y., R.S.H., J.M.-S., J.D.F. and T.W.T. wrote the manuscript and generated the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Sections 1 (1.1–1.4), 2, 3 (3.1, 3.2.1–3.2.7, 3.3.1–3.3.4, 3.4.1 and 3.4.2), 4 (4.1–4.4, 4.4.1-4.4.5, 4.5 and 4.6), 5 (5.1, 5.2, 5.2.1–5.2.4, 5.3–5.5), 6, 7 (7.1–.7.5), 8 and 9.
Rights and permissions
About this article
Cite this article
Marshall, J.R., Yao, P., Montgomery, S.L. et al. Screening and characterization of a diverse panel of metagenomic imine reductases for biocatalytic reductive amination. Nat. Chem. 13, 140–148 (2021). https://doi.org/10.1038/s41557-020-00606-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-020-00606-w
This article is cited by
-
Using enzymes to tame nitrogen-centred radicals for enantioselective hydroamination
Nature Chemistry (2023)
-
Chemoenzymatic approaches for exploring structure–activity relationship studies of bioactive natural products
Nature Synthesis (2023)
-
Actinomycetes-derived imine reductases with a preference towards bulky amine substrates
Communications Chemistry (2022)
-
A growth selection system for the directed evolution of amine-forming or converting enzymes
Nature Communications (2022)
-
Multifunctional biocatalyst for conjugate reduction and reductive amination
Nature (2022)