Abstract
The prolyl hydroxylation of hypoxia-inducible factor 1α (HIF-1α) mediated by the EGLN–pVHL pathway represents a classic signalling mechanism that mediates cellular adaptation under hypoxia. Here we identify RIPK1, a known regulator of cell death mediated by tumour necrosis factor receptor 1 (TNFR1), as a target of EGLN1–pVHL. Prolyl hydroxylation of RIPK1 mediated by EGLN1 promotes the binding of RIPK1 with pVHL to suppress its activation under normoxic conditions. Prolonged hypoxia promotes the activation of RIPK1 kinase by modulating its proline hydroxylation, independent of the TNFα–TNFR1 pathway. As such, inhibiting proline hydroxylation of RIPK1 promotes RIPK1 activation to trigger cell death and inflammation. Hepatocyte-specific Vhl deficiency promoted RIPK1-dependent apoptosis to mediate liver pathology. Our findings illustrate a key role of the EGLN–pVHL pathway in suppressing RIPK1 activation under normoxic conditions to promote cell survival and a model by which hypoxia promotes RIPK1 activation through modulating its proline hydroxylation to mediate cell death and inflammation in human diseases, independent of TNFR1.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Coordinates and structure factors have been deposited in the PDB under the accession code 7CJB. The structure of VCB proteins in complex with a peptide (residue 557–574 with Pro564 hydroxylated) of HIF-1α has been deposited in the PDB under the accession code 4AJY. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
References
Eltzschig, H. K., Bratton, D. L. & Colgan, S. P. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat. Rev. Drug Discov. 13, 852–869 (2014).
Eltzschig, H. K. & Eckle, T. Ischemia and reperfusion—from mechanism to translation. Nat. Med. 17, 1391–1401 (2011).
Pasparakis, M. & Vandenabeele, P. Necroptosis and its role in inflammation. Nature 517, 311–320 (2015).
Yuan, J., Amin, P. & Ofengeim, D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 20, 19–33 (2019).
Xu, D. et al. TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491.e19 (2018).
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012).
Wang, H. et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell 54, 133–146 (2014).
Green, D. R. The coming decade of cell death research: five riddles. Cell 177, 1094–1107 (2019).
Degterev, A. et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1, 112–119 (2005).
Linkermann, A. et al. Two independent pathways of regulated necrosis mediate ischemia-reperfusion injury. Proc. Natl Acad. Sci. USA 110, 12024–12029 (2013).
Adameova, A. et al. Evidence of necroptosis in hearts subjected to various forms of ischemic insults. Can. J. Physiol. Pharmacol. 95, 1163–1169 (2017).
Rosenbaum, D. M. et al. Necroptosis, a novel form of caspase-independent cell death, contributes to neuronal damage in a retinal ischemia-reperfusion injury model. J. Neurosci. Res. 88, 1569–1576 (2010).
Naito, M. G. et al. Sequential activation of necroptosis and apoptosis cooperates to mediate vascular and neural pathology in stroke. Proc. Natl Acad. Sci. USA 117, 4959–4970 (2020).
Crifo, B. et al. Hydroxylase inhibition selectively induces cell death in monocytes. J. Immunol. 202, 1521–1530 (2019).
Polykratis, A. et al. Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 193, 1539–1543 (2014).
Degterev, A. et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008).
Ofengeim, D. et al. Activation of necroptosis in multiple sclerosis. Cell Rep. 10, 1836–1849 (2015).
Zhang, D. W. et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 (2009).
Cho, Y. S. et al. Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009).
He, S. et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137, 1100–1111 (2009).
Bertrand, M. J. et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 (2008).
Stroka, D. M. et al. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 15, 2445–2453 (2001).
Jain, I. H. et al. Hypoxia as a therapy for mitochondrial disease. Science 352, 54–61 (2016).
Vergadi, E. et al. Early macrophage recruitment and alternative activation are critical for the later development of hypoxia-induced pulmonary hypertension. Circulation 123, 1986–1995 (2011).
Florentin, J. et al. Inflammatory macrophage expansion in pulmonary hypertension depends upon mobilization of blood-borne monocytes. J. Immunol. 200, 3612–3625 (2018).
Yu, Y. A. et al. Nonclassical monocytes sense hypoxia, regulate pulmonary vascular remodeling, and promote pulmonary hypertension. J. Immunol. 204, 1474–1485 (2020).
Lee, P., Chandel, N. S. & Simon, M. C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 21, 268–283 (2020).
Wu, K. et al. Stabilization of HIF-1α by FG-4592 promotes functional recovery and neural protection in experimental spinal cord injury. Brain Res. 1632, 19–26 (2016).
Shivaraju, M. et al. Airway stem cells sense hypoxia and differentiate into protective solitary neuroendocrine cells. Science 371, 52–57 (2021).
Long, G. et al. Antianemia drug roxadustat (FG-4592) protects against doxorubicin-induced cardiotoxicity by targeting antiapoptotic and antioxidative pathways. Front. Pharm. 11, 1191 (2020).
Salnikow, K. et al. Depletion of intracellular ascorbate by the carcinogenic metals nickel and cobalt results in the induction of hypoxic stress. J. Biol. Chem. 279, 40337–40344 (2004).
Wang, G. L. & Semenza, G. L. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: implications for models of hypoxia signal transduction. Blood 82, 3610–3615 (1993).
Duan, L. J., Takeda, K. & Fong, G. H. Hematological, hepatic, and retinal phenotypes in mice deficient for prolyl hydroxylase domain proteins in the liver. Am. J. Pathol. 184, 1240–1250 (2014).
Samson, A. L. et al. A toolbox for imaging RIPK1, RIPK3, and MLKL in mouse and human cells. Cell Death Differ. 28, 2126–2144 (2021).
Xie, T. et al. Structural basis of RIP1 inhibition by necrostatins. Structure 21, 493–499 (2013).
Zheng, X. et al. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by blocking its interaction with the USP9x deubiquitinase. Genes Dev. 28, 1429–1444 (2014).
Zhu, K. et al. Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression. Cell Death Dis. 9, 500 (2018).
Jaakkola, P. et al. Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Ivan, M. et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 (2001).
Yu, F., White, S. B., Zhao, Q. & Lee, F. S. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl Acad. Sci. USA 98, 9630–9635 (2001).
Chen, Z. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005).
Min, J. H. et al. Structure of an HIF-1α–pVHL complex: hydroxyproline recognition in signaling. Science 296, 1886–1889 (2002).
Hon, W. C. et al. Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature 417, 975–978 (2002).
Roe, J. S. et al. p53 stabilization and transactivation by a von Hippel–Lindau protein. Mol. Cell 22, 395–405 (2006).
Rankin, E. B. et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol. Cell. Biol. 29, 4527–4538 (2009).
Krishna-Subramanian, S. et al. RIPK1 and death receptor signaling drive biliary damage and early liver tumorigenesis in mice with chronic hepatobiliary injury. Cell Death Differ. 26, 2710–2726 (2019).
Dara, L., Liu, Z. X. & Kaplowitz, N. Questions and controversies: the role of necroptosis in liver disease. Cell Death Discov. 2, 16089 (2016).
Masson, N., Willam, C., Maxwell, P. H., Pugh, C. W. & Ratcliffe, P. J. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 20, 5197–5206 (2001).
Eltzschig, H. K. & Carmeliet, P. Hypoxia and inflammation. N. Engl. J. Med. 364, 656–665 (2011).
Yuan, X. et al. Targeting hypoxia signaling for perioperative organ injury. Anesth. Analg. 126, 308–321 (2018).
Rosenberger, P. et al. Hypoxia-inducible factor-dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nat. Immunol. 10, 195–202 (2009).
Guo, J. et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science 353, 929–932 (2016).
Chowdhury, R. et al. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure 17, 981–989 (2009).
Acknowledgements
We thank M. Kelliher (University of Massachusetts) and M. Pasparakis (University of Cologne) for providing the Ripk1D138N mice. This work was supported in part by National Institutes of Health grants (R35CA253027 and CA177910 to W.W.). The work of D.X. was supported in part by grants from the National Key R&D Program of China (2022ZD0213200). J.Y. was supported in part by the National Natural Science Foundation of China (82188101, 21837004, 91849204 and 92049303). D.X. and J.Y. were supported in part by the Shanghai Municipal Science and Technology Major Project (2019SHZDZX02), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB39030200 to J.Y. and XDB39030600 to D.X.) and Shanghai Key Laboratory of Aging Studies (19DZ2260400). The work of J.L. was supported by a grant from the National Natural Science Foundation of China (32071297). The work of Y.-R.L. was supported in part by grants from the Career Development Award, Academia Sinica Taiwan (AS-CDA-110-L07 to Y.-R.L) and Ministry of Science and Technology Taiwan (110-2320-B-001-029-MY2 to Y.-R.L). We thank W.W. and J.Y. laboratory members for critical reading of the manuscript, as well as members of the W.W., J.Y. and W.G.K. laboratories for helpful discussions.
Author information
Authors and Affiliations
Contributions
T.Z. was the key contributor in designing and conducting the majority of the experiments. T.Z., J.Y. and W.W. conceived of and directed the project and wrote the manuscript. D.X., J.L. and M.W. conducted the key experiments. L.-J.D., M. Liu, H.M., Y.Z., H.W., Yaru Wang, M. Lv, Z.Z., J.H., R.G., X.X., H.L., E.E., Yingnan Wang, W.Y., F.D., D.G., C.J., X.D., H.I., P.Y., J.W., M.B., G.L. and Z.T. conducted some of the experiments. J.M., G.S., G.-H.F., L.P., A.E.K., Y.-R.L. and W.G.K. edited the manuscript. L.S. directed the Ripk1P196A knock-in mouse model study. T.Z., Y.Z. and G.S. evaluated the liver sections from the different mice.
Corresponding authors
Ethics declarations
Competing interests
W.W. is a co-founder and consultant for Rekindle Therapeutics. G.S. is paid consultant for Cyta Therapeutics, DURECT, Evive, Merck, Pfizer, Surrozen, Terra Firma, Pandion Therapeutics, Labcorp, Glympse Bio, Satellite Bio and Zomagen. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Holger Eltzschig and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Hypoxia induces RIPK1 activity.
(a-d) Primary MEFs of indicated genotypes were exposed to 1% O2 for different times. Cell death was measured by PI uptake assay. Data are mean ± s.d. of n = 3 biological independent samples. Two-way ANOVA (a, d). Quantified values for western blot images for Fig. 1b. Data are presented as mean ± s.e.m. of n = 3 independent experiments (b). (e-g) L929 (e, f) and HT-29 (g) cells were exposed to 1% O2 in the presence or absence of Nec-1s. Cell death was measured by PI uptake assay. Data are mean ± s.d. of n = 3 biological independent samples. Two-way ANOVA. Nec-1s, 25 μM (e, g). (h-p) HT-29 (h, i), Jurkat (j, k, l), THP-1 (m, n), primary fibroblasts (o, p) were exposed to 1% O2 for 48 h. Cell death was measured by PI uptake assay. Data are mean ± s.d. of n = 3 biological independent samples. Unpaired two-tailed t-test (j, m, o). (q-s) Immortalized (q) and primary (r, s) WT or Ripk1D138N MEFs were exposed to different oxygen concentrations for 48 h. Cell death was measured by PI uptake assay. Data are mean ± s.d. of n = 3 biological independent samples. Two-way ANOVA (q, r). (t-z) MEFs of indicated genotypes (t, u, v, x, y, z) and primary MEFs treated with or without SM164 (50 nM) (w) were exposed to 1% O2 for the indicated times. Cell death was measured by PI uptake assay. Data are mean ± s.d. of n = 3 biological independent samples, unpaired two-tailed t-test (t, u, x). Cell lysates were immunoblotted with the indicated antibodies(c,f,h, i, k, l, n, p,s,v, w, y, z). Results are representative of three independent experiments (c,f,h, i, k, l, n, p,s,v, w, y, z).
Extended Data Fig. 2 Hypoxia promotes RIPK1 activity in vivo.
(a-i) Mice of indicated genotypes were challenged to hypoxia for 72 h. Histological analysis on brain sections (n = 10) (a), immunostaining for p-RIPK1(S166) on brain sections (n = 10) (b), brain and liver sections (n = 10) (c), qRT-PCR analysis of the mRNA expression of the indicated cytokines and chemokines from brains (n = 6). Data are mean ± s.e.m. One-way ANOVA, post hoc Dunnett’s test (d), TUNEL assays and immunostaining for Cleaved caspase-3 (CC3) on liver sections (n = 6). Scale bars, 100 μm. DAPI for nuclei. Microscopic quantification of TUNEL and CC3 positive cells (right). Data are mean ± s.e.m. Unpaired two-tailed t-test (e), serum ALT and AST levels (f), Bilirubin levels (g) were measured from wild type mice (n = 6). Data are mean ± s.e.m. Unpaired two-tailed t-test, immunostaining for p-RIPK1(S166) (h), CD68 and Ly6G (i) on lung sections (n = 6). Scale bars, 100 μm. DAPI for nuclei. Microscopic quantification of p-S166, CD68 and Ly6G positive cells (right). Data are mean ± s.e.m. One-way ANOVA, post hoc Dunnett’s test.
Extended Data Fig. 3 Inhibition of EGLN1 promotes RIPK1 activation.
(a-b) 293 T cells were transfected with the indicated expression vectors. The lysates were immunoprecipitated with anti-GST, and the immunocomplexes were analysed by immunoblotting using anti-HA (a) or anti-Flag (b). Results are representative of three independent experiments. (c-m) MEFs (c, d, e, f, g, j, k), HT-29 cells (h, l, m), L929 cells (i) were treated with CoCl2 (c-d), DFO (e-f), FG-4592 (g, h, j, k, l, m), 10 μM necrosulfonamide (NSA) for 48 h (m) with indicated concentrations or times as indicated. The levels of indicated proteins were determined by immunoblotting, Results are representative of three independent experiments (c, d, e, f, h, i, j). Cell death was measured by PI uptake assay. Data are mean ± s.d. of n = 3 biological independent samples. Unpaired two-tailed t-test (g, k, l, m). (n-r) The livers from indicated genotypes at the age of 6 weeks. The levels of Eglns were determined by immunoblotting (n = 3), Serum ALT, AST and Bilirubin levels (n = 8), Unpaired two-tailed t-test (o), Masson’s trichrome or Sirius Red staining on liver sections (p) (n = 4), immunostaining for proinflammatory cytokine S100A8 (q), CD11b and Ly-6) (r) on liver sections (n = 4). Microscopic quantification of S100A8, CD11b and Ly-6G positive cells (right). Data are means ± s.e.m. One-way ANOVA, post hoc Dunnett’s test. DAPI for nuclei. Scale bar, 100 μm.
Extended Data Fig. 4 Inhibition of Eglns activity caused liver damage.
(a-h) Co-immunostaining for p-RIPK1(S166) and HNF4α (a), α-SMA (b), VE-Cadherin (c), and CD45 (d), co-immunostaining for CC3 and HNF4α (e), α-SMA (f), VE-Cadherin (g), and CD45 (h) on liver sections from Egln1fl/fl::Egln2fl/fl::Egln3fl/fl and Egln1fl/fl::Egln2fl/fl::Egln3fl/fl::AlbCre littermate mice at the age of 6 weeks (n = 4). DAPI for nuclei. Scale bar, 100 μm. (i) A schematic representation of FG-4592 induction of RIPK1 activation and inflammation. (j-l) Mice of indicated genotypes at the age of 6-8 weeks were intraperitoneally injected with vehicle control or FG-4592 (25 mg/kg, 50 mg/kg or 250 mg/kg body weight) every 24 h for two days (n = 5) (j). WT mice were treated with or without Nec-1s for 7 days, and then were intraperitoneally injected with vehicle control or FG-4592 (250 mg/kg body weight) with or without Nec-1s every 24 h for another two days (n = 6) (k), qRT-PCR analysis of the mRNA expression of the indicated cytokines and chemokines (n = 6 for the Vehicle, n = 8 for FG-4592 or FG-4592+Nec-1s) (l). TUNEL assays and immunostaining for p-RIPK1(S166) on liver sections. DAPI for nuclei. Microscopic quantification of TUNEL and p-RIPK1(S166) positive cells (right). Data are means ± s.e.m. One-way ANOVA, post hoc Dunnett’s test. Data are mean ± s.e.m. One-way ANOVA, post hoc Dunnett’s test.
Extended Data Fig. 5 EGLN1 hydroxylates RIPK1 to suppress RIPK1 activity.
(a-b) 293 T cells were transfected with expression vectors of Flag-RIPK1. Cell lysates were immunoprecipitated with anti-Flag antibody, and the immunocomplexes were analysed by mass spectrometry analysis. MS/MS spectrum to demonstrate the identified hydroxylated RIPK1 peptides at Pro195. The covered amino acids are coloured in yellow, and hydroxylated Proline amino acids are coloured in green. (c) The sequence alignment of human RIPK1 (Homo sapiens), rat RIPK1 (Rattus norvegicus) and murine RIPK1 (Mus musculus) using Clustal Omega is shown around the P195 residue. (d) MEFs cells were infected with expression vectors of Flag-mRIPK1 for 24 h, and then treated with the EGLNs inhibitor CoCl2 (400 μM) for another 16 h. The cell lysates were immunoprecipitated with anti-Flag antibody, and the immunocomplexes were analysed by immunoblotting using anti-pan-hydroxylation antibody. Results are representative of three independent experiments. (e-f) Mass spectrometry analysis detects the presence of proline hydroxylation event at the RIPK1 Pro195 site in the reaction products of in vitro hydroxylation assays with recombinant His-EglN1, and HIF1α peptides were used as positive controls (e). Recombinant RIPK1 kinase domain (1-312) purified from insect cells (f). (g) HT-29 cells were infected with expression vectors of WT or mutant Flag-RIPK1 for 24 h. The cell lysates were immunoprecipitated with anti-Flag antibody, and the immunocomplexes were analysed by western blotting using the anti-p195-OH RIPK1 antibody. Results are representative of three independent experiments. (h) Immunostaining for RIPK1 in primary WT or Ripk1P196A MEFs. DAPI for nuclei. Scale bar, 100 μm. Results are representative of three independent experiments. (i) 293 T cells were treated with or without FG4592 (50 μM) for 24 h. The cell lysates were immunoprecipitated with anti-RIPK1 antibody, and the immunocomplexes were analysed by immunoblotting using the anti-p195-OH RIPK1 antibody. Results are representative of three independent experiments.
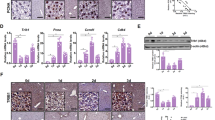
Extended Data Fig. 6 RIPK1 P196A mutation promotes cell death and inflammation in vivo.
(a) schematic diagram of the murine Ripk1 locus and the Ripk1-P196A allele. (b-c) Tail genotyping for the Ripk1P196A mutant mice. Expected results: Ripk1WT/WT = 730 bp, Ripk1WT/P196A = 730 bp, 500 bp and 230 bp, Ripk1P196A/P196A = 500 bp and 230 bp. Results are representative of four independent experiments (b). (d) Numbers of offspring from intercrossing Ripk1P196A/WT parents. (e) Representative images of female WT and Ripk1P196A/P196A mutant mice at 16 weeks of age. (f) Plot of weight changes of the female WT and Ripk1P196A/P196A mutant mice at 4-22 weeks of age (WT, n = 14, and Ripk1P196A/P196A mutant, n = 16). Data are means ± s.e.m. Two-way ANOVA. (g) The indicated tissue weights of Ripk1P196A/P196A mutant mice and the control mice at 16-18 weeks of age (n = 7). Data are means ± s.e.m., Unpaired two-tailed t-test. (h-i) qRT-PCR analysis of the mRNA expression of the indicated cytokines and chemokines in RNA isolated from total small intestine (h) and liver (i) from 16-20-week-old mice with the indicated genotypes (n = 8). Data are means ± s.e.m., Unpaired two-tailed t-test.
Extended Data Fig. 7 Treatment with EGLNs inhibitors promotes RIPK1 activation.
(a-e) TUNEL assays (n = 5) (a), immunostaining for CC3 (n = 5) (b), serum ALT and AST levels (n = 7) (c), serum Bilirubin levels (n = 7) (d) from Ripk1P196A/P196A and control mice. DAPI for nuclei. Scale bar, 100 μm. Microscopic quantification of TUNEL, CC3 positive cells (right). Data are means ± s.e.m., Unpaired two-tailed t-test. qRT-PCR analysis of the mRNA expression of the indicated cytokines and chemokines from Ripk1P196A/P196A and control mice (n = 10) or Ripk1P196A/P196A treated with Nec-1s (n = 5) for 7 days. Data are means ± s.e.m., One-way ANOVA, post hoc Dunnett’s test (e). (f) A schematic representation of FG-4592 induction of RIPK1 activation and inflammation. (g-h) qRT-PCR analysis of the mRNA expression of HIF pathway downstream targets Glut1 and Vegf-a from livers (n = 6) (g), TUNEL assays and immunostaining for p-RIPK1(S166) on intestine sections (n = 5) (h) of 16-20-week-old mice with the indicated genotypes treated with or without FG-4592 at the dose of 25 mg/kg body weight for 7 days. Data are means ± s.e.m., One-way ANOVA, post hoc Dunnett’s test. DAPI for nuclei. Microscopic quantification of TUNEL and p-RIPK1(S166) positive cells (bottom). (i-l) Immunostaining for p-RIPK1(S166) (i), TUNEL assays (j), Serum ALT and AST levels (k), ELISA analyses of CCL2 and IL6 protein expression (l) from Ripk1P196A/P196A and control mice treated with or without FG-4592 at the dose of 250 mg/kg body weight for 2 days (n = 8). DAPI for nuclei. Scale bar, 100 μm. Microscopic quantification of p-RIPK1(S166) and TUNEL positive cells (right). Data are means ± s.e.m., Unpaired two-tailed t-test.
Extended Data Fig. 8 pVHL suppresses RIPK1 activity.
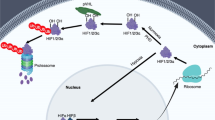
(a-c) Cell lysates from primary human fibroblasts (a), RIPK1-/- HT-29 (b,c) cells infected with indicated expression vectors with (b) or without (c) DFO (200 μM) were immunoprecipitated. The immunocomplexes were immunoblotted. Data are mean ± s.e.m. of n = 3 independent experiments (c). (d-f) Cell lysates from 293 T cells transfected with indicated expression vectors with (e) or without (d) 10 μM Nec-1s, immunoprecipitated using anti-Flag (e), the reaction products of in vitro hydroxylation assays (f), incubated with or without 100 μM ATP, 50 μM Nec-1s or purified recombinant VHL and hydroxylated HIF1α peptide at 30 °C for 30 min. The samples were analysed by immunoblotting. (g) A sequence alignment of indicated proteins using the Clustal Omega program. (h) A schematic representation of various biotinylated synthetic RIPK1-derived peptides. (i) Cell lysates from 293 T cells transfected with indicated expression vectors for 24 h, were incubated with RIPK1-derived peptides for 4 h. The immunocomplexes were analysed by immunoblotting. (j) Coomassie staining of the bacterially purified recombinant pVHL/Elongin-C/Elongin-B complex. (k-l) A structural overlay of pVHL/Elongin-C/Elongin-B bound to either HIF1α or RIPK1. (m) Details of interaction between pVHL and HIF-1α (PDB: 4AJY). (n) 293 T cells were transfected with indicated expression vectors for 24 h. The levels of indicated proteins were determined by immunoblotting. (o-q) Cell lysates from Vhl KO MEFs (o), Vhl KO HT-29 cells (p), Cul2 knockdown HT-29 cells (q) were immunoblotted. (r-t) Cell lysates from VHL KO 293 T cells (r), MEFs (s), and 786-O cells (t) were immunoprecipitated with anti-RIPK1 antibody, and immunoblotted. (u-v) Vhl KO MEFs reconstituted with indicated expression vectors exposed to hypoxia. Cell death was measured by PI uptake assay. Data are mean ± s.d. of n = 3 biological independent samples. Two-way ANOVA, post hoc Bonferroni’s test (u). Results are representative of three independent experiments. (a-f, i, j,n-t, v).
Extended Data Fig. 9 Loss of hydroxylation of RIPK1 sensitized cell to cell death.
(a-e) Primary WT or Ripk1P196A MEFs were treated with TNFα/Smac (a,c), TNFα/Smac/zVAD.fmk (b,d,e) in the presence or absence of Nec-1s. Cell death were measured by PI uptake assay (a,b). The levels of indicated proteins were determined by immunoblotting. Results are representative of three independent experiments (c, d). Cell lysates were immunoprecipitated with anti-RIPK3 antibody, and the immunocomplexes were analysed by western blotting using anti-RIPK1 antibody. Results are representative of three independent experiments (e). Nec-1s, 20 μM; TNF, 10 ng/mL; SM164, 50 nM; zVAD.fmk, 20 μM. Data are mean ± s.d. of n = 3 biological independent samples. Two-way ANOVA. (f-k) MEFs of indicated genotypes were treated with TNFα/CHX/zVAD.fmk (f, g), TNFα/Smac/zVAD.fmk (h, i), TNFα/5z7 (j, k) in the presence or absence of Nec-1s (h, j, k) or FG-4592 (i). Cell death were measured by PI uptake assay (f, h, i) and by ATP-Glo assay, Data are mean ± s.d. of n = 3 biological independent samples. Two-way ANOVA, post hoc Bonferroni’s test (j). The levels of pS166-RIPK1 and RIPK1 were determined by immunoblotting (g, k). Data are mean ± s.d. of n = 3 biological independent samples. One-way ANOVA, post hoc Dunnett’s test (f), Data are mean ± s.e.m. of n = 3 independent experiments (g, k). Data are mean ± s.d. of n = 3 biological independent samples. Unpaired two-tailed t-test (h), Data are mean ± s.d. of n = 3 biological independent samples. Two-way ANOVA, post hoc Dunnett’s test (i). TNFα, 10 ng/mL; zVAD.fmk, 20 μM; CHX, 1 μg/mL; SM164, 50 nM; FG-4592, 200 μM; 5z7, 200 nM; (l-m) Vhl KO MEFs treated with TNFα/SM-164/zVAD (l) and VHL-deficient 786-O cells treated with T/5z7/S (m) in the presence or absence of Nec-1s as indicated for different time followed by cell viability analyses using ATP-Glo assay. Data are means ± s.d. of n = 3 biological independent samples. Two-way ANOVA. Compounds used: Nec-1s, 10 μM; TNFα, 10 ng/mL; SM-164, 50 nM; zVAD.fmk, 20 μM; 5z7, 500 nM.
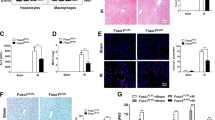
Extended Data Fig. 10 VHL deletion in liver promotes RIPK1 activation and cell death.
(a) Representative images of mice of indicated genotypes at 5 weeks of age. (b-f) Histological analysis (b), TUNEL assays (c, e), immunostaining for HNF4α(e) and p-RIPK1(S166) (f), microscopic quantification of TUNEL positive cells from (c) and p-RIPK1(S166) (d, f) on liver sections from mice of indicated genotypes (n = 5). DAPI for nuclei. Scale bar, 100 μm. (g-i) Liver sections from mice of indicated genotypes at 5 weeks of age, subjected to Oil Red O staining (n = 3) (g), immunostaining for cleaved caspase-3 (CC3) (n = 5) (h), microscopic quantification of CC3 positive cells from (h) (i). DAPI for nuclei. Scale bar, 100 μm. (j) qRT-PCR analysis of the mRNA expression of the downstream of HIF pathway targets from liver sections of mice of indicated genotypes at the age of 6-8 weeks (n = 3). (k-l) Representative images of spleens (k) and livers (l) of mice of indicated genotypes. Spleen weights (k)(Vhlf/f, n = 17, Vhlf/f;Alb-Cre, n = 13; Vhlf/f;Alb-Cre;Ripk1D138N/D138N, n = 8) and Liver weights (l) (Vhlf/f, n = 17, Vhlf/f;Alb-Cre, n = 13; Vhlf/f;Alb-Cre;Ripk1D138N/D138N, n = 10). (m-n) Representative images of liver sections stained with the F4/80 antibodies (m) and Masson’s trichrome or Sirius Red staining (n) from 4-week-old WT, Vhl-LKO or Vhl-LKO;Ripk1D138N/D138N mice (n = 3), Scale bars, 100 μm. (o) Body weight of the control WT (n = 17), Vhl-LKO (n = 17), Vhl-LKO;Ripk1D138N/D138N (n = 8) mice. (p-r) Immunostaining for p-RIPK1(S166) (p), TUNEL assays (q), and qRT-PCR analysis of the mRNA expression of the indicated cytokines and chemokines (r) on liver sections from mice of indicated genotypes treated with Nec-1s for 7 days (n = 6). DAPI for nuclei. Scale bar, 100 μm. Microscopic quantification of p-RIPK1(S166) and TUNEL positive (right). Data are means ± s.e.m., Unpaired two-tailed t-test (d,i). Data are means ± s.e.m., One-way ANOVA, post hoc Dunnett’s test(j,k,l,n,o,p,q,r).
Supplementary information
Supplementary Information
Full Worldwide PDB X-ray structure validation report.
Supplementary Tables
Supplementary Table 1. Data collection and refinement statistics. Supplementary Table 2. List of primers for quantitative reverse transcription PCR analysis of the indicated genes.
Source data
Source Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 8
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 9
Unprocessed western blots and/or gels.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, T., Xu, D., Liu, J. et al. Prolonged hypoxia alleviates prolyl hydroxylation-mediated suppression of RIPK1 to promote necroptosis and inflammation. Nat Cell Biol 25, 950–962 (2023). https://doi.org/10.1038/s41556-023-01170-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-023-01170-4
This article is cited by
-
Hypoxia-stabilized RIPK1 promotes cell death
Nature Cell Biology (2023)
-
A guide to cell death pathways
Nature Reviews Molecular Cell Biology (2023)