Abstract
While the acquisition of cellular plasticity in adult stem cells is essential for rapid regeneration after tissue injury, little is known about the underlying mechanisms governing this process. Our data reveal the coordination of airway progenitor differentiation plasticity by inflammatory signals during alveolar regeneration. Following damage, interleukin-1β (IL-1β) signalling-dependent modulation of Jag1 and Jag2 expression in ciliated cells results in the inhibition of Notch signalling in secretory cells, which drives the reprogramming and acquisition of differentiation plasticity. We identify the transcription factor Fosl2 (also known as Fra2) for secretory cell fate conversion to alveolar type 2 cells that retain the distinct genetic and epigenetic signatures of secretory lineages. We also reveal that human secretory cells positive for KDR (also known as FLK-1) display a conserved capacity to generate alveolar type 2 cells via Notch inhibition. Our results demonstrate the functional role of an IL-1β–Notch–Fosl2 axis in the fate decision of secretory cells during injury repair, proposing a potential therapeutic target for human lung alveolar regeneration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The ATAC-seq, RNA-seq and scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession codes GSE153677 (RNA-seq and ATACC-seq for organoids) and GSE154218 (scRNA-seq). Previous published sequencing data that were re-analysed here are available under accession codes EGAS00001004344 (ref. 44), GSE135893 (ref. 43) and GSE144553 (ref. 31). Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Fuchs, E., Tumbar, T. & Guasch, G. Socializing with the neighbors: stem cells and their niche. Cell 116, 769–778 (2004).
Blanpain, C. & Fuchs, E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014).
Butler, J. P. et al. Evidence for adult lung growth in humans. N. Engl. J. Med. 367, 244–247 (2012).
Hogan, B. L. et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014).
Barkauskas, C. E. et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013).
Rock, J. R. et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl Acad. Sci. USA 108, E1475–E1483 (2011).
Adamson, I. Y. & Bowden, D. H. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab. Invest. 30, 35–42 (1974).
Kathiriya, J. J., Brumwell, A. N., Jackson, J. R., Tang, X. & Chapman, H. A. Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell 26, 346–358.e4 (2020).
Vaughan, A. E. et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625 (2015).
Salwig, I. et al. Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. EMBO J. https://doi.org/10.15252/embj.2019102099 (2019).
Liu, Q. et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar–duct junction. Nat. Genet. 51, 728–738 (2019).
Guha, A., Deshpande, A., Jain, A., Sebastiani, P. & Cardoso, W. V. Uroplakin 3a+ cells are a distinctive population of epithelial progenitors that contribute to airway maintenance and post-injury repair. Cell Rep. 19, 246–254 (2017).
Rawlins, E. L. et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534 (2009).
Miller, A. J. et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 14, 518–540 (2019).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Nikolic, M. Z. et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife https://doi.org/10.7554/eLife.26575 (2017).
Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. https://doi.org/10.15252/embj.2018100300 (2019).
Lee, J. H. et al. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170, 1149–1163.e12 (2017).
Shiraishi, K. et al. In vitro expansion of endogenous human alveolar epithelial type II cells in fibroblast-free spheroid culture. Biochem. Biophys. Res. Commun. 515, 579–585 (2019).
Weiner, A. I. et al. Mesenchyme-free expansion and transplantation of adult alveolar progenitor cells: steps toward cell-based regenerative therapies. NPJ Regen. Med. 4, 17 (2019).
Katsura, H. et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27, 890–904.e8 (2020).
Youk, J. et al. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell 27, 905–919.e10 (2020).
Maillard, I. et al. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 203, 2239–2245 (2006).
Pardo-Saganta, A. et al. Parent stem cells can serve as niches for their daughter cells. Nature 523, 597–601 (2015).
Lafkas, D. et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 528, 127–131 (2015).
Morimoto, M. et al. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of clara versus ciliated cell fate. J. Cell Sci. 123, 213–224 (2010).
Yum, M. K. et al. Tracing oncogene-driven remodelling of the intestinal stem cell niche. Nature 594, 442–447 (2021).
You, P. et al. Jagged-1–HES-1 signaling inhibits the differentiation of TH17 cells via ROR gammat. J. Biol. Regul. Homeost. Agents 27, 79–93 (2013).
Tsao, P. N. et al. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136, 2297–2307 (2009).
Choi, J. et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e7 (2020).
Rawlins, E. L., Ostrowski, L. E., Randell, S. H. & Hogan, B. L. Lung development and repair: contribution of the ciliated lineage. Proc. Natl Acad. Sci. USA 104, 410–417 (2007).
Kotton, D. N. & Morrisey, E. E. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat. Med. 20, 822–832 (2014).
Kimura, S. et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10, 60–69 (1996).
Martis, P. C. et al. C/EBPα is required for lung maturation at birth. Development 133, 1155–1164 (2006).
Holla, V. R., Mann, J. R., Shi, Q. & DuBois, R. N. Prostaglandin E2 regulates the nuclear receptor NR4A2 in colorectal cancer. J. Biol. Chem. 281, 2676–2682 (2006).
Ke, N. et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 64, 8208–8212 (2004).
Koppula, P., Zhang, Y., Zhuang, L. & Gan, B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. 38, 12 (2018).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Lee, J. H. et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4–NFATc1–thrombospondin-1 axis. Cell 156, 440–455 (2014).
Chen, H. et al. Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells 30, 1948–1960 (2012).
Chernaya, O., Shinin, V., Liu, Y. & Minshall, R. D. Behavioral heterogeneity of adult mouse lung epithelial progenitor cells. Stem Cells Dev. 23, 2744–2757 (2014).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Geng, Y. et al. Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proc. Natl Acad. Sci. USA 108, 7058–7063 (2011).
Weaver, M., Yingling, J. M., Dunn, N. R., Bellusci, S. & Hogan, B. L. Bmp signaling regulates proximal–distal differentiation of endoderm in mouse lung development. Development 126, 4005–4015 (1999).
Rouillard, A. D. et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge aboutgenes and proteins. Database https://doi.org/10.1093/database/baw100 (2016).
Zepp, J. A. et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170, 1134–1148.e10 (2017).
Chilosi, M. et al. Aberrant Wnt/β-catenin pathway activation in idiopathic pulmonary fibrosis. Am. J. Pathol. 162, 1495–1502 (2003).
Jensen-Taubman, S. M., Steinberg, S. M. & Linnoila, R. I. Bronchiolization of the alveoli in lung cancer: pathology, patterns of differentiation and oncogene expression. Int. J. Cancer 75, 489–496 (1998).
Xu, Y. et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1, e90558 (2016).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Robson, M. J. et al. Generation and characterization of mice expressing a conditional allele of the interleukin-1 receptor type 1. PLoS ONE 11, e0150068 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Gaspar, J. M. NGmerge: merging paired-end reads via novel empirically-derived models of sequencing errors. BMC Bioinformatics 19, 536 (2018).
Shao, Z., Zhang, Y., Yuan, G. C., Orkin, S. H. & Waxman, D. J. MAnorm: a robust model for quantitative comparison of ChIP-seq data sets. Genome Biol. 13, R16 (2012).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Consortium, G. O. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Bult, C. J. et al. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 47, D801–D806 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Acknowledgements
We would like to thank E. Rawlins (Gurdon Institute, University of Cambridge, UK) for sharing Scgb1a1-CreERTM, Foxj1-CreERT2 and Rosa26R-CAG-fGFP mouse lines; I. Pshenichnaya (histology), M. Paramor (next-generation sequencing library), P. Humphreys (imaging), A. Riddell (flow cytometry), S. McCallum (flow cytometry, Cambridge NIHR BRC Cell Phenotyping Hub), K. Kania (single-cell sequencing at Cancer Research UK), W. Mansfield (transgenic mice) and Cambridge Stem Cell Institute core facilities for technical assistance; J.-E. Park (Welcome Sanger Institute, UK) and S. Han (Gurdon institute, UK) for helpful discussions on the scRNA-seq analysis; Y. Ahn (DGIST, Korea) for technical assistance for the shRNA experiment; and all Lee Lab members for helpful discussion. This work was supported by Wellcome and the Royal Society (107633/Z/15/Z) and European Research Council Starting Grant (679411). J.K. is supported by R01GM112722 from the National Institute of General Medical Sciences and a Preterm Birth Research Grant from the Burroughs Wellcome Fund. S.M.J. is funded by the Medical Research Council UKRMP2, a Cancer Research UK Programme grant and the Lung Cancer Centre of Excellence. B.D.S. acknowledges funding from the Royal Society E.P. Abraham Research Professorship (RP/R1/180165) and the Wellcome Trust (098357/Z/12/Z).
Author information
Authors and Affiliations
Contributions
J.C., Y.J.J., J.K. and J.-H.L. designed the experiments, interpreted the data and wrote the manuscript. J.C. performed most of the experiments and data analyses. Y.J.J. performed and analysed the bulk RNA-seq and ATAC-seq data. C.D. designed and performed the lineage-tracing analysis of Red2-NotchN1ICD mice and provided valuable comments on the manuscript. E.I. generated the Sftpc-IRES-DTR-P2A-dsRed targeting vector. K.V.E. performed the isolation of human lung tissue. J.-H.L. generated the Sftpc-IRES-DTR-P2A-dsRed mouse line. B.-K.K. helped with the generation of the Sftpc-IRES-DTR-P2A-dsRed targeting vector and shared the Red2-NotchN1ICD mouse line. B.D.S. helped with the study of Red2-NotchN1ICD lineage-tracing analysis. H.H. and S.M.J. provided human lung tissue samples.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Tien Peng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
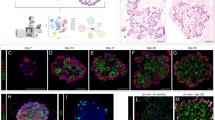
Extended Data Fig. 1 Establishment of feeder-free organoids derived from AT2 cells.
a, Schematics of experimental design for isolation of Sftpc lineage-labelled AT2 cells at indicated time points after tamoxifen treatment. b, Representative bright-field images of organoids derived from lineage-labelled Tomato+Sftpc+ cells in indicated conditions; complete medium with WNT3A, RSPO1 (R-spondin 1), EGF, FGF7, FGF10, and NOG (Noggin), withdrawal of indicated factors (−FGF10, −FGF7, or −WNT3A/RSPO1). Scale bar, 2,000μm. c, d, Statistical quantification of colony forming efficiency (c, n = 5) and passaging numbers (d, n = 5) of organoids. Each individual dot represents individual biological replicate and data are presented as mean and s.e.m. Statistical analysis was performed using two-tailed unpaired Student’s t test; n.s; not significant. e, Representative serial bright-field images of a lung organoid growing originated from single Tomato+Sftpc+ cells at the indicated time points. Magnifications: X20 (day 4 and 7), X10 (day10 and 13), and X4 (day 16, 20, and 30). Scale bars, 400μm. f, A representative immunofluorescence (IF) image of organoids derived from Tomato+Sftpc+ cells at the first passage under feeder-free condition with complete culture medium. SPC (for AT2 cells, red), Pdpn (for AT1 cells, green), and DAPI (blue). Scale bars, 50μm. g, Representative bright-field images of organoids under feeder-free condition with complete culture medium at passage 5. Insets (left) show high-power view (right). Scale bars, 2,000μm. h, Representative IF images of mixed organoids cultured in complete medium at passage >5. SPC (red), Hopx (white), and DAPI (blue). Scale bars, 50μm.
Extended Data Fig. 2 Interference of Notch activity in secretory cell-derived organoids (SCOs) enhances the differentiation into AT2 lineages.
a, Representative IF images showing nuclear localisation of intracellular domain of Notch 1 (N1ICD) in SCOs at passage 5 with or without DAPT (20 μM). CC10 (red), N1ICD (white), and DAPI (blue). Scale bars, 50μm. b, qPCR analysis of gene expression in control (control KD) or Rbpj knock-downed (Rbpj KD) organoids. Each individual dot represents one individual experiment (n = 4 biological replicates) and data are presented as mean ± s.e.m. Statistical analysis was performed using two-tailed unpaired Student’s t test; P-values are indicated in the figure. c, Representative IF images of control KD or Rbpj KD organoids: CC10 (green), SPC (red), and DAPI (blue). Scale bars, 50μm. d, Representative IF images showing Hes1 expression in lineage-labelled secretory cells after tamoxifen treatment in the indicated genotype: Tomato (for Scgb1a1 lineage, red), Hes1 (white), and DAPI (blue). Scale bars, 50μm. e, qPCR analysis of Hes1 expression in isolated EpCAM+Tomato+. Each individual dot represents one individual experiment (n = 2 individual biological replicate). f, Representative IF images showing the derivation of lineage-labelled Act-Tub+ ciliated cells in PBS- or bleomycin-treated lungs of Scgb1a1-CreERTM/+;R26RtdTomato/+ or dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ mice. Tomato (for Scgb1a1 lineage, red), Acetylated-Tubulin (Act-Tub, green), and DAPI (blue). Scale bar, 50μm. g, Statistical quantification of lineage-labelled ciliated cells: n = 19 sections (PBS), n = 20 sections (Bleo), pooled from 2 mice for Scgb1a1-CreERTM/+;R26RtdTomato/+; n = 20 sections (PBS) and n = 20 sections (Bleo), pooled from 2 mice for dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+).
Extended Data Fig. 3 Organoid co-culture shows enhanced differentiation of secretory cells into AT2 cells by downregulation of Notch activity.
a, Experimental design for isolation of lineage-labelled secretory cells from Scgb1a1-CreERTM/+;R26RtdTomato/+ and dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ mice at indicated time points after tamoxifen treatment. b, Representative fluorescent images of organoids derived from control or dnMAML-expressing lineage-labelled secretory cells. 5,000 cells of lineage-labelled secretory cells (Tomato+EpCAM+CD45−CD31−) isolated from Scgb1a1-CreERTM/+;R26RtdTomato/+ or dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ mouse lungs were co-cultured with stromal cells with 1:5 ratio for 14 days. Arrowheads point to cystic airway-like organoids. Scale bar, 2,000μm. c, Statistical quantification of colony forming efficiency of organoids. Each individual dot represents one biological replicate (n = 5 for control, n = 6 for dnMAMLflox/+) and data are presented as mean and s.e.m. Statistical analysis was performed using two-tailed unpaired Student’s t test; n.s; not significant. CFE; Colony forming efficiency units. d, Representative IF images of three distinctive types of organoids derived from control secretory cells; Airway organoids (CC10+SPC−; denoted as 1), Alveolar organoids (CC10−SPC+; denoted as 2), and Mixed organoids (CC10+SPC+; denoted as 3). CC10 (for secretory cells, red), SPC (for AT2 cells, white), and DAPI (blue). Insets (1, 2 and 3) show high-power view. Scale bars, 50μm and 10μm (in high-power view). e, Representative IF images of alveolar organoids derived from dnMAML-expressing secretory cells. CC10 (for secretory cells, red), SPC (for AT2 cells, white), and DAPI (blue). Insets show high-power view. Scale bars, 50μm (10μm in high-power view). f, Quantification of each organoid types derived from Scgb1a1-CreERTM/+;R26RtdTomato/+ or dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ mice in (d and e); Airway organoids (CC10+SPC−; red), Alveolar organoids (CC10−SPC+; blue), and Mixed organoids (CC10+SPC+; grey). Data are presented as mean and s.e.m (n = 3 biological replicates). Statistical analysis was performed using two-tailed unpaired Student’s t test; *p=0.0358. g, qPCR analysis of markers for secretory (Scgb1a1, Scgb3a2, Gabrp, Cldn10, and Cyp2f2) and AT2 (Sftpc, Etv5, Lamp3, Lpcat1, and Abca3) cells in organoids isolated from Scgb1a1-CreERTM/+;R26RtdTomato/+ (black bars) and dnMAMLflox/+;Scgb1a1-CreERTM/+; R26RtdTomato/+ mice (white bars). Data are presented as mean ± s.e.m (n = 3 biological replicates). Statistical analysis was performed using two-tailed unpaired Student’s t test; P-values are indicated in the figure.
Extended Data Fig. 4 Pharmacological inhibition of Notch activity by DAPT treatment enhances the differentiation of secretory cells into AT2 cells during injury repair.
a, Experimental design of lineage-tracing analysis of Scgb1a1-CreERTM/+;R26RtdTomato/+ mice after bleomycin injury. DMSO or DAPT (50 mg/kg body weight) was administrated via intraperitoneally as indicated time points. b, Representative IF images showing the increased Scgb1a1+ lineage-labelled Act-Tub+ ciliated cells in DAPT-treated mouse lungs at day 21 post bleomycin injury compared to DMSO control mice. Tomato (for Scgb1a1 lineage, red), Acetylated Tubulin (Act-Tub, white), and DAPI (blue). Scale bar, 50μm. c,d, Representative IF images showing the derivation of Scgb1a1+ lineage-labelled SPC+ AT2 (left) or CC10+ secretory cells (right) in DMSO control (c) or DAPT-treated (d) mice at day 21 post bleomycin injury. Tomato (for Scgb1a1 lineage, red), SPC (white), CC10 (green), and DAPI (blue). Scale bar, 100μm.
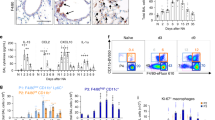
Extended Data Fig. 5 Transplantation of secretory cells, after excluding lineage-labelled AT2 cells, reveals enhanced the differentiation of secretory cells into AT2 cells via Notch inhibition.
a, MHCII expression marks AT2 cells. Representative flow cytometry analysis of MHCII expression in SPC+ AT2 (EpCAM+dsRed+) or non-AT2 (EpCAM+dsRed−) cells from Sftpc-dsRedIRES-DTR reporter mice. Sftpc-IRES-DTR-P2A-dsRed (Sftpc-dsRedIRES-DTR) reporter mouse was used to monitor SPC-expressing AT2 cells based on the expression of dsRed (See Methods). Numbers adjacent to the outlined area indicate the percentage of populations. b, Flow cytometry analysis to exclude lineage-labelled AT2 cells, including CC10+SPC+ dual-positive cells, from dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ or Scgb1a1-CreERTM/+;R26RtdTomato/+ mouse lungs. c-h, Representative IF image of engrafted lineage-labelled secretory cells (EpCAM+Tomato+MHCII−) isolated from Scgb1a1-CreERTM/+;R26RtdTomato/+ (c-e) or dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ (f-h) mouse lungs at day 14 post transplantation. 20,000 cells of EpCAM+Tomato+MHCII− were engrafted into injured lung at day 7 after bleomycin injury. Tomato (for Scgb1a1 lineage, red), SPC (white), and DAPI (blue). Scale bars, 100μm. i, Representative fluorescent images of organoids (left) and statistical quantification of colony forming efficiency of alveolar organoids (right) derived from lineage-labelled AT2 cells isolated from control (Scgb1a1-CreERTM/+;R26RtdTomato/+) or dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ mouse lungs. 5,000 cells of lineage-labelled secretory cells (EpCAM+Tomato+MHCII−) isolated from Scgb1a1-CreERTM/+;R26RtdTomato/+ or dnMAMLflox/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ mice were co-cultured with stromal cells with 1:5 ratio for 14 days. Each individual dot represents one individual experiment (n = 2 biological replicates). Scale bar, 2,000μm. j, Representative IF images showing Scgb1a1+ lineage-labelled cells in a whole lobe of Scgb1a1-CreERTM/+;Red2-NotchN1ICD/+ mice post tamoxifen treatment: YFP (yellow), RFP (red), pro-SPC (white, left), Scgb1a1 (white, right) and DAPI (blue). Scale bar, 500μm (left), and 100μm (right). Of note, only airway secretory cells were labelled by tamoxifen induction.
Extended Data Fig. 6 scRNA-seq analysis of lung cells from Scgb1a1-CreERTM/+;Red2-NotchN1ICD/+ mice at day 28 post bleomycin injury.
a, Sorting strategy for Scgb1a1 lineage-labelled RFP+ and YFP+ or unlabelled (YFP−RFP−) cells by flow cytometry after bleomycin injury. For unlabelled population (YFP−RFP−), EpCAM−RFP−YFP− (non-epithelial population) and EpCAM+RFP−YFP− (epithelial population) cells were mixed at 1:1 ratio. b, Clusters of cell population after bleomycin injury from 10x Genomics 3′ scRNA-seq analysis visualised by UMAP, assigned by specific colours. c, A heatmap showing gene expression patterns of key markers in each distinctive cell cluster. d, e, Representative IF images showing the expansion of Porcn+ or p57+ cells derivation from RFP+ cells at day 28 post bleomycin injury. RFP (red), CC10 (green), p57 (white, e), Porcn (white, f), and DAPI (blue). Insets (1) show high-power view. Scale bar, 50μm. f, Diffusion map according to diffusion pseudotime (DPT, right) order coloured by samples (left).
Extended Data Fig. 7 Expression of Notch ligands in ciliated cells post alveolar injury. a, Expression of Notch ligand in each distinctive cluster revealed in Extended Data Fig. 6.
The size of the circle represents the fraction of cells expressing the gene and the colour represents the relative expression of each gene. b, UMAP visualisation of the log-transformed (log10(TPM+1)), normalised expression of selected marker genes (Jag1 and Jag2 for Notch ligand expression; Foxj1 for ciliated cells) in distinctive clusters. c, Representative IHC images stained with anti-Jag1 in the distal airway of lungs from Scgb1a1-CreERTM/+;R26RtdTomato/+ mice at day 14 post PBS or bleomycin treatment. Insets (top) show high-power view (bottom). Scale bars, 50μm. d, Sorting strategy for Il1r1+ lineage-labelled ciliated (EpCAM+Tomato+CD24high) and AT2 (EpCAM+Tomato+CD24−) cells from Il1r1-CreERT2/+; R26RtdTomato/+ mouse lungs at day 4 post tamoxifen injection. e, qPCR analysis of Foxj1, Cd24, and Sftpc expression in isolated cells in (d). Each individual dot represents one individual experiment (n = 4) and data are presented as mean ± s.e.m. P-values using Student’s t-test are indicated in the figure. f, Representative fluorescent images of organoids derived from Il1r1 lineage-labelled ciliated (Il1r1+CD24high) or AT2 (Il1r1+CD24−) cells. Ciliated cells are unable to form organoids. Scale bars, 2,000μm. g, Representative IF images showing intact ciliated cells in the lungs of Il1r1flox/flox;Foxj1-CreERT2/+;Scgb1a1-CreERTM/+;R26RtdTomato/+ mice compared to those in the lungs of control Il1r1flox/flox; Scgb1a1-CreERTM/+;R26RtdTomato/+ mice: Tomato (for Scgb1a1 or Foxj1 lineage, red), Acetylated Tubulin (Act-Tub, white), E-Cadherin (E-Cad, green), and DAPI (blue). Scale bars, 50μm. h, Representative IF images showing a small number of Scgb1a1 lineage-labelled AT2 cells at day 28 post PBS treatment in Scgb1a1-CreERTM/+;Il1r1flox/flox;R26RtdTomato/+ or Foxj1-CreERT2/+;Scgb1a1-CreERTM/+;Il1r1flox/flox;R26RtdTomato/+ mice. Tomato (for Scgb1a1 lineage, red), SPC (white), and DAPI (blue). Arrowheads point to lineage-labelled SPC+ AT2 cells. Scale bar, 100μm. i, Representative fluorescent images of organoids derived from lineage-labelled secretory cells of Scgb1a1-CreERTM/+;R26RtdTomato/+ lungs. Organoids were cultured with PBS or IL-1β for 14 days. Arrows point to cystic airway-like organoids. Scale bar, 2,000μm. j, Flow cytometry analysis of secretory cell-derived AT2 cells isolated from Scgb1a1-CreERTM/+;R26RfGFP/+;Sftpc-dsRedIRES-DTR/+ mouse lungs after PBS (left) or bleomycin (right) treatment.
Extended Data Fig. 8 Differential genetic and epigenetic characteristics between secretory cells and secretory-derived AT2 (sAT2) cells.
a, A spearman correlation map plotted by using ATAC-seq replicates obtained from secretory and sAT2 cells. b, A bar graph showing the chromosomal distribution of secretory-specific and sAT2-specific cis-regulatory elements, mapped by ATAC-seq in secretory and sAT2 cells across the mouse genome. c, The distribution of ATAC-seq peak signals in secretory and sAT2 cells near the centre of secretory-specific, shared, and sAT2-specific open regions. d, Bar graphs showing enriched GO terms of the genes nearby secretory and sAT2-specific ATAC-seq peaks, respectively. e, Signal track images showing open chromatin regions nearby markers for secretory (Gabrp and Cyp2f2) and AT2 cells (Sftpc and Lyz2) in secretory (blue) and sAT2 cells (red). f, A heatmap showing differentially expressed transcription factors (TFs) in secretory-derived organoids with or without DAPT treatment. g, qPCR analysis of the markers for secretory (Scgb1a1 and Gabrp) and AT2 cells (Sftpc, Etv5, and Lamp3) in secretory cell-derived organoids (SCOs) with or without DAPT treatment after knockdown of control (white), Etv5 (black), or Cebpa (red).Each individual dot represents one individual experiment (n = 4) and data are presented as mean ± s.e.m. h, qPCR analysis to confirm the knockdown efficiency of the genes that are used for indicated constructs in SCOs. Each individual dot represents one individual biological experiment (n = 4 for knockdown of Fosl2, Rpbjl, Etv5, and C/ebpa, n = 3 for knockdown of Srebf2) and data are presented as mean ± s.e.m. Statistical analysis was performed using two-tailed unpaired Student’s t test; P-values are indicated in the figure.
Extended Data Fig. 9 Genetic and epigenetic differences of sAT2 cells compared to rAT2 cells.
a, Experimental design for isolation of secretory-derived (sAT2, GFP+dsRed+) and resident AT2 cells (non-lineage-labelled AT2, rAT2, GFP−dsRed+) using Scgb1a1-CreERTM/+;R26RfGFP/+;Sftpc-dsRedIRES-DTR/+ mice. Specific time points for tamoxifen injection and isolation are indicated. b, Representative flow cytometry analysis for isolation of sAT2 (GFP+dsRed+) and rAT2 (GFP−dsRed+) cells. c, Heatmaps showing open chromatin regions specific in sAT2 cells and shared open chromatin regions between sAT2 cells and rAT2 cells (left). A pie chart presenting the proportion of sAT2-specific (blue) and shared regions (grey) (right). d, A bar graph showing the chromosomal distribution of sAT2-specific ATAC-seq peaks mapped in (c). e, A bar graph showing enriched GO terms of the genes nearby sAT2-specific peaks. f, GSEA with gene sets representing the negative regulation of intrinsic apoptosis signalling pathway in scRNA-seq data of sAT2 and rAT2 shown in Fig. 6a. g, A heatmap showing the expression patterns of the genes belonging to the negative regulation of apoptosis signalling pathway in sAT2 and rAT2 cells monitored by scRNA-seq (Fig. 6a). h, Signal track image showing open regions for Nr4a2, anti-apoptotic pathway marker, in secretory (blue), sAT2 (red), and rAT2 cells (green). i, Representative IF images of organoids derived from rAT2 or sAT2 cells. SPC (for AT2 cells, red), HOPX (for AT1 cells, white), and DAPI (blue). Scale bars, 50μm. j, k, Quantification of the frequency of AT2 (SPC+) or AT1 (HOPX+) cells (b) and the ratio of AT1/AT2 cells (c) in rAT2- or sAT2-derived organoids. (n = 23 organoids, pooled from 2 mice for rAT2; n = 30 organoids, pooled from 2 mice for sAT2). l, Flow cytometry analysis of Scgb1a1+ lineage-labelled AT2 cells isolated from Scgb1a1-CreERTM/+;R26RfGFP/+;Sftpc-dsRedIRES-DTR/+ mouse lungs at 3 months after bleomycin injury. m, qPCR analysis of the genes in isolated secretory, rAT2, or sAT2 cells. Data are presented as mean ± s.e.m (n = 5 biological replicates for all experimental groups).
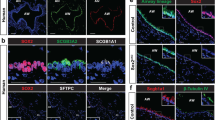
Extended Data Fig. 10 Identification of KDR/Flk-1 as a surface marker of secretory cells in both mouse and human lungs.
a, UMAP visualisation of normalised expression of Kdr/Flk-1. b, A signal track image showing open regions nearby Kdr mapped in secretory (blue) and sAT2 cells (red). c, Experimental design for isolation of CC10+SPC− secretory cells, CC10+SPC+ dual-positive cells including Bronchioalveolar stem cells (BASCs), and CC10−SPC+ AT2 cells from Scgb1a1-CreERTM/+;R26RfGFP/+;Sftpc-dsRedIRES-DTR/+ mice. d, Representative flow cytometry analysis (left) for secretory (GFP+dsRed−), BASCs (GFP+dsRed+), and AT2 (GFP−dsRed+) cells. e, A bar graph showing MFI (mean fluorescence of intensity) of KDR expression. Experiment performed twice. f,g, UMAP (f) or t-SNE (g) visualisation of the normalised expression of JAG1, HES1, and KDR from the dataset of Habermann et al.45(f) or Travaglini et al.46(g). h,i, Representative IF images of secretory cell-derived organoids treated with DMSO or DAPT (20 μM). CC10 (white), Acetylated Tubulin (Act-Tub, green, h), KRT5 (red, i), and DAPI (blue). Scale bar, 50μm. j, Quantification of the frequency of Act-Tub+ ciliated cells or KRT5+ basal cells. Data are presented as mean ± s.e.m. (n = 8 (Control) and n = 7 (DAPT) organoids, pooled from 3 mice for Act-TUB+ cells; n = 5 (Control) and n = 8 (DAPT) organoids, pooled from 3 mice for KRT5+ cells). Statistical analysis was performed using two-tailed unpaired Student’s t test; n.s.=not significance. k, Notch inhibition coordinates the cell fate decision of secretory cells into AT2 cells in two stages: (1) Reprogramming; loss of secretory cell identity by Notch inhibition in response to IL-1β signaling and (2) AT2 cell conversion; acquisition of transcriptional programmes for AT2 cell differentiation vis the transcription regulator Fosl2. l, Representative IF images of secretory organoids treated with DMSO (control) or DAPT (20 μM) in the presence (middle) or withdrawals of Wnt-inducing factors (right). Organoids were cultured in the medium supplemented with Wnt3a/Rspo1 for 7 days and then DAPT was treated for further 7 days with or without Wnt3a/Rspo1. CC10 (for secretory cells, green), Acetylated Tubulin (for ciliated cells, Act-Tub, red), SPC (for AT2 cells, white), and DAPI (blue). Scale bars, 50μm. m, The expression of IL-1β, Wnt5a, and Wnt7b at indicated time points after bleomycin injury. Data were from our previous study31.
Supplementary information
Supplementary Tables
Supplementary Table 1: Sequences of shRNA for KD. Supplementary Table 2: Sequences for qPCR analysis.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Choi, J., Jang, Y.J., Dabrowska, C. et al. Release of Notch activity coordinated by IL-1β signalling confers differentiation plasticity of airway progenitors via Fosl2 during alveolar regeneration. Nat Cell Biol 23, 953–966 (2021). https://doi.org/10.1038/s41556-021-00742-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00742-6
This article is cited by
-
Rho kinase inhibitor Y-27632 downregulates IL-1β expression in mice with experimental autoimmune myocarditis
Scientific Reports (2024)
-
A single-cell atlas of in vitro multiculture systems uncovers the in vivo lineage trajectory and cell state in the human lung
Experimental & Molecular Medicine (2023)
-
Abnormal respiratory progenitors in fibrotic lung injury
Stem Cell Research & Therapy (2022)
-
Function of epithelial stem cell in the repair of alveolar injury
Stem Cell Research & Therapy (2022)
-
Human distal airways contain a multipotent secretory cell that can regenerate alveoli
Nature (2022)