Abstract
In many cancers, high proliferation rates correlate with elevation of rRNA and tRNA levels, and nucleolar hypertrophy. However, the underlying mechanisms linking increased nucleolar transcription and tumorigenesis are only minimally understood. Here we show that IMP dehydrogenase-2 (IMPDH2), the rate-limiting enzyme for de novo guanine nucleotide biosynthesis, is overexpressed in the highly lethal brain cancer glioblastoma. This leads to increased rRNA and tRNA synthesis, stabilization of the nucleolar GTP-binding protein nucleostemin, and enlarged, malformed nucleoli. Pharmacological or genetic inactivation of IMPDH2 in glioblastoma reverses these effects and inhibits cell proliferation, whereas untransformed glia cells are unaffected by similar IMPDH2 perturbations. Impairment of IMPDH2 activity triggers nucleolar stress and growth arrest of glioblastoma cells even in the absence of functional p53. Our results reveal that upregulation of IMPDH2 is a prerequisite for the occurance of aberrant nucleolar function and increased anabolic processes in glioblastoma, which constitutes a primary event in gliomagenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Metabolomics data (Supplementary Tables 1–6), information on patient cohorts (Supplementary Table 7) and microarray data (Supplementary Table 9) are available in the Supplementary Information. Microarray data that support the findings of this study have also been deposited in the GEO under the accession code GSE124727. Gene expression data for Supplementary Fig. 2b,c are available from the GlioVis website (http://gliovis.bioinfo.cnio.es/). Probe ID and gene symbols for Supplementary Fig. 2a are provided in Supplementary Table 8. Previously published microarray and sequencing data that were re-analysed here are available under accession code GSE45874. The primer list used in this study is provided in Supplementary Table 10. Source data for Figs. 1–7 and Supplementary Figs. 1–7 have been provided in Supplementary Table 11. The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Pianese, G. Beitrag zur Histologie und Aetiologie der Carcinoma. Histologische und experimentelle Untersuchungen. Beitr. Pathol. Anat. Allg. Pathol. 142, 1–193 (1896).
Ruggero, D. Revisiting the nucleolus: from marker to dynamic integrator of cancer signaling. Sci. Signal. 5, pe38 (2012).
Hara, A. et al. Correlation between nucleolar organizer region staining and Ki-67 immunostaining in human gliomas. Surg. Neurol. 33, 320–324 (1990).
Busch, H., Byvoet, P. & Smetana, K. The nucleolus of the cancer cell: a review. Cancer Res. 23, 313–339 (1963).
Tsai, R. Y. L. & Pederson, T. Connecting the nucleolus to the cell cycle and human disease. FASEB J. 28, 3290–3296 (2014).
Grummt, I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma 122, 487–497 (2013).
Woese, C. R. & Forro, J. R. Correlations between ribonucleic acid and deoxyribonucleic acid metabolism during spore germination. J. Bacteriol. 80, 811–817 (1960).
de Srulijes, L. K., Israeli, E. & Barzilai, D. Brief communication: RNA:DNA ratios in human tumors. J. Natl Cancer Inst. 58, 769–770 (1977).
Thompson, M., Haeusler, R. A., Good, P. D. & Engelke, D. R. Nucleolar clustering of dispersed tRNA genes. Science 302, 1399–1401 (2003).
Drygin, D., Rice, W. G. & Grummt, I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 50, 131–156 (2010).
Traut, T. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 (1994).
Allsop, J. & Watts, R. W. Purine synthesis and salvage in brain and liver. Adv. Exp. Med. Biol. 165(Pt B), 21–26 (1984).
Seegmiller, J. E., Rosenbloom, F. M. & Kelley, W. N. Enzyme defect associated with a sex-linked human neurological disorder and excessive purine synthesis. Science 155, 1682–1684 (1967).
Nyhan, W. L. Behavior in the Lesch–Nyhan syndrome. J. Autism Child. Schizophr. 6, 235–252 (1976).
Gerweck, L. E. & Wakimoto, H. At the crossroads of cancer stem cells, radiation biology, and radiation oncology. Cancer Res. 76, 994–998 (2016).
Cloughesy, T. F., Cavenee, W. K. & Mischel, P. S. Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. Mech. Dis. 9, 1–25 (2014).
Nakano, I. Stem cell signature in glioblastoma: therapeutic development for a moving target. J. Neurosurg. 14, 324–330 (2014).
Lathia, J. D., Mack, S. C., Mulkearns-Hubert, E. E., Valentim, C. L. L. & Rich, J. N. Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217 (2015).
Hosios, A. M. & Vander Heiden, M. G. The redox requirements of proliferating mammalian cells. J. Biol. Chem. 293, 7490–7498 (2018).
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Carr, S., Papp, E., Wu, J. & Natsumeda, Y. Characterization of human type I and type II IMP dehydrogenases. J. Biol. Chem. 268, 27286 (1993).
Hager, P. W., Collart, F. R., Huberman, E. & Mitchell, B. S. Recombinant human inosine monophosphate dehydrogenase type I and type II proteins. Purification and characterization of inhibitor binding. Biochem. Pharmacol. 49, 1323–1329 (1995).
Hedstrom, L. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 109, 2903–2928 (2009).
Jackson, R. C., Weber, G. & Morris, H. P. IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature 256, 331–333 (1975).
Carter, S. B. et al. Mycophenolic acid: an anti-cancer compound with unusual properties. Nature 223, 848–850 (1969).
Williams, R. H., Lively, D. H., DeLong, D. C., Cline, J. C. & Sweeny, M. J. Mycophenolic acid: antiviral and antitumor properties. J. Antibiot. 21, 463–464 (1968).
Majd, N. et al. A review of the potential utility of mycophenolate mofetil as a cancer therapeutic. J. Cancer Res. 2014, 1–12 (2014).
Valvezan, A. J. et al. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability. Cancer Cell 32, 624–638.e5 (2017).
Wang, X. et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci. 20, 661–673 (2017).
Huang, F. et al. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab. 28, 369–382.e5 (2018).
Horbinski, C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 125, 621–636 (2013).
Bentley, R. Mycophenolic acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 100, 3801–3826 (2000).
Sintchak, M. D. & Nimmesgern, E. The structure of inosine 5′-monophosphate dehydrogenase and the design of novel inhibitors. Immunopharmacology 47, 163–184 (2000).
Yokota, S. Mizoribine: mode of action and effects in clinical use. Pediatr. Int. 44, 196–198 (2002).
Dudzinska, W., Lubkowska, A., Dolegowska, B., Safranow, K. & Jakubowska, K. Adenine, guanine and pyridine nucleotides in blood during physical exercise and restitution in healthy subjects. Eur. J. Appl. Physiol. 110, 1155–1162 (2010).
Jové, M., Portero-Otín, M., Naudí, A., Ferrer, I. & Pamplona, R. Metabolomics of human brain aging and age-related neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 73, 640–657 (2014).
Drygin, D. et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 71, 1418 (2011).
Peltonen, K. et al. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 25, 77–90 (2014).
Pelletier, J., Thomas, G. & Volarevic, S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer 18, 51–63 (2018).
Grummt, I. & Grummt, F. Control of nucleolar RNA synthesis by the intracellular pool sizes of ATP and GTP. Cell 7, 447–453 (1976).
Huang, M., Ji, Y., Itahana, K., Zhang, Y. & Mitchell, B. Guanine nucleotide depletion inhibits pre-ribosomal RNA synthesis and causes nucleolar disruption. Leuk. Res. 32, 131–141 (2008).
Nguyen, L. X. T. et al. Regulation of ribosomal RNA synthesis in T cells: requirement for GTP and Ebp1. Blood 125, 2519–2529 (2015).
Thompson, D. M. & Parker, R. Stressing out over tRNA cleavage. Cell 138, 215–219 (2009).
Tsai, R. Y. L. & McKay, R. D. G. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Devel. 16, 2991–3003 (2002).
Lo, D., Dai, M. S., Sun, X. X., Zeng, S. X. & Lu, H. Ubiquitin- and MDM2 E3 ligase-independent proteasomal turnover of nucleostemin in response to GTP depletion. J. Biol. Chem. 287, 10013–10020 (2012).
Zhai, W. & Comai, L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20, 5930–5938 (2000).
Liebhaber, S. A., Wolf, S. & Schlessinger, D. Differences in rRNA metabolism of primary and SV40-transformed human fibroblasts. Cell 13, 121–127 (1978).
Gu, J. J. Induction of apoptosis in IL-3-dependent hematopoietic cell lines by guanine nucleotide depletion. Blood 101, 4958–4965 (2003).
Park, J. et al. Mycophenolic acid inhibits platelet-derived growth factor-induced reactive oxygen species and mitogen-activated protein kinase activation in rat vascular smooth muscle cells. Am. J. Transplant. 4, 1982–1990 (2004).
Emmanuel, N. et al. Purine nucleotide availability regulates mTORC1 activity through the Rheb GTPase. Cell Rep. 19, 2665–2680 (2017).
Eugui, E. M., Almquist, S. J., Muller, C. D. & Allison, A. C. Lymphocyte-selective cytostatic and immunosuppressive effects of mycophenolic acid in vitro: role of deoxyguanosine nucleotide depletion. Scand. J. Immunol. 33, 161–173 (1991).
Liu, Y.-C. et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE 3, e2722 (2008).
Mourad, M. et al. Correlation of mycophenolic acid pharmacokinetic parameters with side effects in kidney transplant patients treated with mycophenolate mofetil. Clin. Chem. 47, 88–94 (2001).
Nguyen, T., Park, J. Y., Scudiere, J. R. & Montgomery, E. Mycophenolic acid (cellcept and myofortic) induced injury of the upper GI tract. Am. J. Surg. Pathol. 33, 1355–1363 (2009).
Chen, L. & Pankiewicz, K. W. Recent development of IMP dehydrogenase inhibitors for the treatment of cancer. Curr. Opin. Drug Discov. Devel. 10, 403–412 (2007).
Sampetrean, O. et al. Invasion precedes tumor mass formation in a malignant brain tumor model of genetically modified neural stem cells. Neoplasia 13, 784–791 (2011).
Kwon, C.-H. et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 68, 3286–3294 (2008).
Chow, L. M. L. et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19, 305–316 (2011).
Steed, T. C. et al. Differential localization of glioblastoma subtype: implications on glioblastoma pathogenesis. Oncotarget 7, 24899–24907 (2016).
Ozawa, T. et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26, 288–300 (2014).
Sumita, K. et al. The lipid kinase PI5P4Kβ is an intracellular GTP sensor for metabolism and tumorigenesis. Mol. Cell 61, 187–198 (2016).
Krycer, J. R. et al. Dynamic metabolomics reveals that insulin primes the adipocyte for glucose metabolism. Cell Rep. 21, 3536–3547 (2017).
Soga, T. et al. Metabolomic profiling of anionic metabolites by capillary electrophoresis mass spectrometry. Anal. Chem. 81, 6165–6174 (2009).
Hirayama, A. et al. Effects of processing and storage conditions on charged metabolomic profiles in blood. Electrophoresis 36, 2148–2155 (2015).
Minami, N. et al. Organotypic brain explant culture as a drug evaluation system for malignant brain tumors. Cancer Med. 6, 2635–2645 (2017).
Sugiura, Y., Honda, K. & Suematsu, M. Development of an imaging mass spectrometry technique for visualizing localized cellular signaling mediators in tissues. Mass Spectrom. 4, A0040–A0040 (2015).
Shiota, M. et al. Gold-nanofève surface-enhanced Raman spectroscopy visualizes hypotaurine as a robust anti-oxidant consumed in cancer survival. Nat. Commun. 9, 1561 (2018).
Bowman, R. L., Wang, Q., Carro, A., Verhaak, R. G. W. & Squatrito, M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncology 19, 139–141 (2017).
Andersen, J. S. et al. Nucleolar proteome dynamics. Nature 433, 77–83 (2005).
Marin-Valencia, I. et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15, 827–837 (2012).
Acknowledgements
We thank the members of the Sasaki Lab, C. Mercer, D. Plas, T. Cunnigham, R. Hatakeyama, K. Ito, A. Kawakami, K. Kono, T. Nakamura, K. Sasaki, R. Kamata, Y. Hirota, T. Senda, K. Takeuchi, Y. Zheng, K. Hazeki, K. Nigorikawa and the Izayoi Society for feedback. We thank E. Dobbs for excellent editing. S. Kofuji is supported, in part, by Home for Innovative Researchers and Academic Knowledge Users (HIRAKU), JSPS KAKENHI grant number JP18K07233 and the Kanae Foundation. R.K. is supported by Grant-in-Aid for JSPS Fellows Grant Number JP17J01882. H.Y. was supported by the Uehara Memorial Foundation. K.S. was supported, in part, by the American Association of Neurological Surgeons. Work in the laboratory of D.A. was funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust (FC001033). R.J.D. is supported by grants from the NIH (R35CA220449). The MS imaging infrastructure was supported by the JST ERATO Suematsu Gas Biology Project (M. Suematsu, to March 2015). H.B. was supported by the Thuringian state programme ProExzellenz (RegenerAging—FSU-I-03/14) of the Thuringian Ministry for Research. The work is supported in part by a UC College of Medicine Research Innovation grant, an MTP UC-Brain Tumor Center grant, a MERF grant, a Marlene Harris Ride Cincinnati grant, an ABTA Discovery grant, a B*Cured research grant, an Ohio Cancer Research grant, R21NS100077 and R01NS089815 (A.T.S.).
Author information
Authors and Affiliations
Contributions
S. Kofuji and A.T.S. conceived and designed the study. S. Kofuji, A.O.E., Y.I., H.Y., D.Y., K.S., K.W., L.L., H. Shimada, A.M., S. Morioka, Y.B., M.A., V.L.F., A.R., J.K., J.T., T.D., K.K., M. Sasaki and M.M. performed the experiments. A.H., S.I. and T.S. performed metabolomics analyses. R.K. performed bioinformatics analyses. O.S., N. Minami, H. Saya helped with ex vivo experiments. Y.S. and M. Suematsu performed MS imaging. M.W., N.S. and W.Y. helped with IHC analysis. S. Kitahara helped with electron microscopy. L.M.L.C., T.M., G.L., S. Mareninov, T.O., Y.K., H.W., E.C.H., W.H.Y., C.H., I.N., R.J.D. and R.M.B. provided samples. N.O., K.O., T.W.-D., N. Majd, E.P.S., D.A., H.W., P.S.M. and I.G. helped with manuscript editing. S. Kofuji, H.B. and A.T.S wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.J.D. is an advisor for Agios Pharmaceuticals. P.S.M. is a cofounder, has equity and serves as a consultant for Pretzel Therapeutics, Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 GTP metabolism is reprogrammed in glioblastoma.
a, A schematic diagram of carbon flux of glucose to purine nucleotides. Red and white dots represents 13C and 12C, respectively. M denotes mass of the unlabeled metabolite. Above M+3 labeling of GTP or ATP are indicative of biosynthesized from the fed [U-13C]glucose. b, GTP pool size increases from neural stem cells (NSC) to its isogenic glioma stem cells (GSC). Data are presented as mean+s.d. n=3 biologically independent samples. c, Mass isotopomer analysis of nucleotides in the indicated cells. U87MG, LN229 and primary glia are cultured for 4 h with [U-13C]glucose. Data are presented as an average of 3 biologically independent samples. d, e, Metabolic flux analysis with [U-13C]glucose shows continuous ATP biosynthesis is utilized for NAD+ and SAM production in GBM, primary glia, NSC and GSC. Data are presented as an average of 3 biologically independent samples (d, left and e) and as mean+s.d., n=3 biologically independent samples (d, right). One-way ANOVA. f, Salvage purine nucleosides are more rapidly incorporated in primary glia than U87MG. The guanosine concentration in the medium was quantified by ion chromatography (IC)-MS. Data are presented as mean+s.d. n=3 biologically independent samples. Unpaired two-sided Student’s t-test. g, 15N-guanosine is incorporated into GTP in primary glia. Primary glia cells were treated with 1 μM [U-15N]-guanosine for 4 h and then cellular metabolite were analyzed by IC-MS. Data are presented as mean+s.d. n=3 biologically independent samples. h, Primary glia is more sensitive to 6-thioguanine (6-TG) than GBM cells. The indicated cells were treated with 6-TG and their cell viability were assessed by crystal violet assay. Data are presented as mean+s.d. n=3 biologically independent samples. Unpaired two-sided Student’s t-test. i, j, NSC utilizes more salvage GTP biosynthesis than GSC. NSC and GSC were incubated with [U-13C, 15N]-labeled glutamine for 24 h and cellular metabolites were analyzed by IC-MS. A schematic diagram of nitrogen flux from glutamine to GTP. Data are presented as an average of 3 biologically independent samples (i) and as mean+s.d., n=3 biologically independent samples (j). Unpaired two-sided Student’s t-test.
Supplementary Figure 2 IMPDH2 mRNA is upregulated in murine and human GBM.
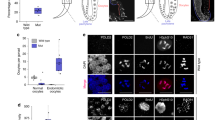
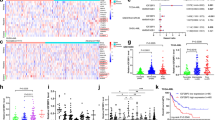
a, Schema showing replication-competent avian sarcoma-leukosis (RCAS)/tv-a system to induce glioma. To investigate the molecular mechanism underlying the metabolic GTP switch, we took advantage of ten genetically different, clinically relevant glioma mice generated by using the RCAS/tv-a system60. In addition to Impdh2, GMP synthetase (Gmps), which converts IMPDH-produced XMP to GMP, is also upregulated. Hprt1, an essential enzyme for salvage GTP biosynthesis, is downregulated. In contrast, adenine phosphoribosyltransferase (Aprt), a critical enzyme for salvage ATP biosynthesis, and AMP deaminase-3 (Ampd3), which converts AMP to IMP, are upregulated. Together with the Impdh2 upregulation in GBM, these results suggest that a systematic change in salvage pathways occur in GBM to decrease GTP salvage and increases ATP salvage production. b, IMPDH2 mRNA is upregulated in human GBM patients. Analyses of five different GBM database on the indicated genes are shown. Similar to the results of the above murine glioma transcriptome analysis, IMPDH2, GMPS, and APRT mRNA are is upregulated in GBM, while HPRT1 and AMPD2 are downregulated. IMPDH1 mRNA expression levels in GBM are similar to normal brain tissue with some variety. TCGA/RNA-seq (n=160), Rembrandt (n=537), Gravendeel (n=284), Gill (n=92), Murat (n=84). Boxplots were derived from GlioVis website. Boxplots follow a Turkey style, in which lower and upper hinges correspond to the first and third quartiles based on their R source file for GlioVis (https://github.com/msquatrito/shiny_GlioVis/blob/master/server.R#L1, version Apr 3, 2018). c, Coordinated metabolic reprograming of purine biosynthesis pathway with IMPDH2 in GBM. Correlation analysis of representative genes in purine synthesis pathway. Plots indicate gene expression data from TCGA glioblastoma patients (HG-U133A platform, n=538). Pearson’s correlation coefficient (R) values are shown. P-values (two-sided) were calculated using the t-distribution with n-2 degrees of freedom.
Supplementary Figure 3 IMPDH2 protein levels are upregulated in murine and human GBM.
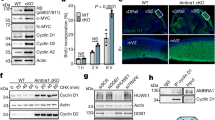
a, Validation of IMPDH1 and IMPDH2 antibodies for their isotype specificity and reactivity in western blot. Of note, upregulation of IMPDH1 in IMPDH2 knockdown cells was observed due to the compensatory mechanism1. n=1 experiment. b, Validation of IMPDH1 and IMPDH2 antibodies for isotype specificity and reactivity in immunohistochemical (IHC) analysis. IMPDH1 and IMPDH2 immuno-staining in paraffin-embedded IMPDH-knocked down and Flag-IMPDH-overexpressing 293T cells. Each IMPDH1 and IMPDH2 antibody selectively recognizes endogenous IMPDH1 and IMPDH2, respectively and does not react with over-expressed IMPDH2 and IMPDH1, respectively. Of note, endogenous IMPDH1 is upregulated upon IMPDH2 knockdown as seen in (a). n=1 experiment. c, IMPDH2 expression in the Gfap-CreER;Ptenfl/fl;Rbfl/fl;Tp53fl/fl;Rbl1-/- mouse brain analyzed by western blotting (upper, n=4 independent animals) and immunohistochemistry (lower, a representative of 8 independent animals). d, No detectable IMPDH1 protein in the Cohort #4. n=1 for grade 2, n=5 for grade 3, n=30 for grade 4. e, Quantification of cellular IMPDH1 and IMPDH2 protein levels show higher IMPDH2 concentration than IMPDH1 in GBM. His-tagged IMPDH1 and IMPDH2 recombinant proteins—for standard—and cell lysates from GBM cells were subjected to SDS-PAGE. After detection of each IMPDH isoform, cellular IMPDH concentrations were calculated using the standard curves generated by the recombinant IMPDHs and cellular volume as described in the method section. n=1 experiment. f, g, IMPDH2 protein levels are much higher than IMPDH1 in GBM. Western blotting analysis using lysates from GBM cell lines, human and mouse GBM stem cells, primary glia and mouse NSC. Non-tagged and Flag-tagged IMPDHs were used as internal controls to compare the expression levels of IMPDH1 and IMPDH2 in GBM cells. n=1 experiment.
Supplementary Figure 4 Requirement of IMPDH activity for cell proliferation and compensatory role of IMPDH1 in IMPDH2 KO cells.
a, The indicated IMPDH KO GBM cells were generated by the CRISPR/Cas9 system as in the method. Western blot shows the upregulation of IMPDH1 in IMPDH2 KO LN229 cells. Compensatory role of IMPDH1 to support cell proliferation of IMPDH2 KO in LN229 and A172, similar to U87MG cells. Cells were maintained with 100 μM guanosine-supplemented media, which was replaced to DMEM/10% dialyzed FBS media 24 h before the assay. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA. n=1 experiment for Western blot analysis. b, c, VX-497, a direct IMPDH inhibitor33, and Mizoribine, a prodrug IMPDH inhibitor, suppress GBM cell proliferations. Unlike MPA and VX-497, mizoribine requires intracellular adenylate kinase to become an active form34. Data are presented as an average of 3 biologically independent samples (b) and mean+s.d. n=3 biologically independent samples (c). Unpaired two-sided Student’s t-test. d, HPRT1 KO U87MG, LN229 and A172 cells can proliferate comparably to parental cells. Western blot analysis verified the depletion of HPRT1 proteins in the transduced cells. The HPRT1 KO U87MG, LN229 and A172 cells were resistant to 6-TG, while the proliferation of their parental cells was significantly decreased above 10 μM 6-TG (upper right). Cells were treated with 6-TG at the indicated concentrations for 72 or 96 h. The HPRT1 KO U87MG, LN229 and A172 cells show comparable proliferation to their parental cells under normal condition (lower right). Proliferation was assessed at 72 or 96 h after plating cells. n=1 experiment for Western blot analysis. Data are presented as mean+s.d. n=6 (U87MG), n=3 (LN229, A172) biologically independent samples. Unpaired two-sided Student’s t-test.
Supplementary Figure 5 IMPDH activity is critical for GBM to promote growth in vivo.
a, Growth of murine GSC in brain tissue is inhibited by MPA in the presence of physiologically relevant guanosine. GFP-expressing murine GSC brain explant slices in immunocompetent mice were cultured with or without 10 μΜ MPA and 1 or 10 μM guanosine. Scale bar indicates 300 μm. Presented data are from 3 out of 4 different mice. Scale bar indicates 300 μm. b, MMF administration suppresses GBM growth. Kinetics of tumor size and mouse weight corresponding to Fig. 4f are shown. Data are presented as mean+s.d. Vehicle (n=3), MMF treatment (n=5). c, Depletion of IMPDH2 gene decreases tumor progression of GBM. Kinetics of tumor size corresponding to Fig. 4g is shown. Data are presented as mean+s.d. from Control (n=6), IMPDH1 KO (n=5), IMPDH2 KO (n=6), IMPDH DKO (n=7) groups. d, Compensatory upregulation of IMPDH1 in IMPDH2 KO in GBM growing in the brain. GFP-tagged luciferase expressing IMPDH2 KO U87MG cells and parental U87MG were implanted in the brain. Bioluminescence imaging showed IMPDH2 KO tumor growth at 3 weeks. The dissected IMPDH2 KO tumor cells from the brain dramatically increased IMPDH1 protein levels. Presented data are from n=2 different animals. Scale bar indicates 200 μm.
Supplementary Figure 6 rRNA is the major sink of newly synthesized GTP in GBM cells.
a, FACS-based cell cycle analysis of U87MG cells shows <20% of cells are in S-phase. Data are representatives from n=2 independent experiments. b, Acute inhibition of GMPS, a downstream enzyme of IMPDH, by Acivicin leads to depletion of GTP within 4 h, phenocopying the effect of MPA. Data are presented as mean+s.d. n=3 biologically independent samples. Unpaired two-sided Student’s t-test. c, Marginal accumulation of dGTP and uric acid, one of the final catabolites of GTP, in U87MG cell as well as in the cultured media. U87MG cells were labeled with [U-13C]glucose for 4 h and polar metabolites were collected and analyzed by CE-MS as in the method. N.D. stands for not detectable. Data are presented as mean+s.d. n=3 biologically independent samples. d, BMH-21 blocks the MPA-dependent depletion of GTP in U87MG and LN229 cells. GTP concentrations were quantified by HPLC in 10 μM MPA (4 h) and 10 μM BMH-21 (5 h including 1 h pretreatment)-treated U87MG and LN229 cells. Data are presented as mean+s.d. n=3 biologically independent samples. One-way ANOVA. e, A detailed schematic diagram of the procedure of SI-MOIRAI.
Supplementary Figure 7 IMPDH inhibition in GBM provokes nucleolar stress response and inhibition of RNA Pol I.
a, Comparison of pre-rRNA levels. n=1 biologically independent sample normalized from three technical replicates. b, IMPDH inhibition by 10 μM MPA for 4 h suppresses tRNA synthesis. Mature transcripts of the indicated genes were analyzed by Q-PCR. Data are presented as mean±s.d. n=4 biologically independent samples. Unpaired two-sided Student’s t-test. c, Effects of IMPDH inhibition by 10 μM MPA in U87MG cells on the p53/p21 pathway. Data are representatives from n=2 independent experiments. d, e, The cells were treated with or without 10 μM MPA and 100 μM guanosine for 24 h, and immunostained by the indicated antibodies and dye. n=1 experiment (d). n=2 independent experiments (e). Scale bar indicates 50 μm (e). f, The indicated GBM cells were treated with MPA for 4 h and 5-fluorouridine for 20 min. Nascent RNA and nucleolus were visualized by anti-BrdU and anti-nucleolin antibodies, respectively. n=1 experiment (A172), n=2 independent experiment (LN229). Scale bar indicates 20 μm. g, Pol I activity at the promoter in GBM cells. The indicated cells were transfected with the rDNA reporter pHrP2-BH and treated with 10 μM MPA for 4 h. The graph represents the level of reporter transcripts normalized to 18S rRNA as determined by Q-PCR. Data are presented as mean+s.e.m. n=5 (U87MG), n=4 (LN229) biologically independent samples. Unpaired two-sided Student’s t-test. h, i, Nucleolar size in LN229 (left), A172 (right) and mImpdh2-reconstituted IMPDH2 KO cells (e). The area of nucleoli from 10 different cells was assessed at 48 h of 10 μM of MPA, 100 μM of guanosine treatment. Data are presented as mean±s.d. One-way ANOVA. Scale bar indicates 50 μm. The results were confirmed in two independent experiments (h). n=1 experiment (i). j, 30 cells/group were assessed from n=3 independent tumors. Arrow heads indicate nucleoli. Data are presented as mean±s.d. **P<0.01, Mann-Whitney test (two-sided). Scale bar indicates 20 μm. k, GSC brain explant slices in immunocompetent mice were cultured with or without 10 μΜ MPA. Data are presented as mean±s.d. 30 cells/group were assessed from n=3 independent samples. Mann-Whitney test (two-sided). Scale bar indicates 20 μm.
Supplementary Figure 8
Unprocessed scans of western blot.
Supplementary information
Supplementary Information
Supplementary Figures 1–8 and Supplementary table titles/legends.
Supplementary Table 1
Metabolites of GBM tissue from PDX.
Supplementary Table 2
Metabolites in primary glia and U87MG cells.
Supplementary Table 3
Metabolites in LN229 cells.
Supplementary Table 4
Metabolites in GBM neurospheres.
Supplementary Table 5
Metabolites in IMPDH-KO U87MG cells.
Supplementary Table 6
Measurement of RNAs in SI-MOIRAI.
Supplementary Table 7
Information on human patient samples.
Supplementary Table 8
Probe IDs and gene symbols used in the gene expression analyses of the RCAS/tv-a system.
Supplementary Table 9
Microarray data and list of ribosome-related genes whose expression changes upon MPA treatment.
Supplementary Table 10
List of primers used in this study.
Supplementary Table 11
Statistics source data.
Rights and permissions
About this article
Cite this article
Kofuji, S., Hirayama, A., Eberhardt, A.O. et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol 21, 1003–1014 (2019). https://doi.org/10.1038/s41556-019-0363-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0363-9