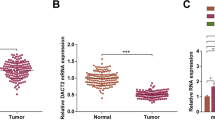
Abstract
Metabolic reprogramming is a hallmark of cancer. Here, we demonstrate that tumour-associated macrophages (TAMs) enhance the aerobic glycolysis and apoptotic resistance of breast cancer cells via the extracellular vesicle (EV) transmission of a myeloid-specific lncRNA, HIF-1α-stabilizing long noncoding RNA (HISLA). Mechanistically, HISLA blocks the interaction of PHD2 and HIF-1α to inhibit the hydroxylation and degradation of HIF-1α. Reciprocally, lactate released from glycolytic tumour cells upregulates HISLA in macrophages, constituting a feed-forward loop between TAMs and tumour cells. Blocking EV-transmitted HISLA inhibits the glycolysis and chemoresistance of breast cancer in vivo. Clinically, HISLA expression in TAMs is associated with glycolysis, poor chemotherapeutic response and shorter survival of patients with breast cancer. Our study highlights the potential of lncRNAs as signal transducers that are transmitted between immune and tumour cells via EVs to promote cancer aerobic glycolysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Microarray and RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE115040 and GSE123474. Source data for Figs. 1–8 and Supplementary Figs. 1–8 have been provided as Statistics Source Data. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
References
DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G. & Thompson, C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 (2008).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Hsu, P. P. & Sabatini, D. M. Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
DeBerardinis, R. J. & Chandel, N. S. Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200 (2016).
Lee, N. & Kim, D. Cancer metabolism: fueling more than just growth. Mol. Cells 39, 847–854 (2016).
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Hensley, C. T. et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016).
Ibrahim-Hashim, A. et al. Defining cancer subpopulations by adaptive strategies rather than molecular properties provides novel insights into intratumoral evolution. Cancer Res. 77, 2242–2254 (2017).
Lloyd, M. C. et al. Darwinian dynamics of intratumoral heterogeneity: not solely random mutations but also variable environmental selection forces. Cancer Res. 76, 3136–3144 (2016).
Semenza, G. L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671 (2013).
Semenza, G. L. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci. 33, 207–214 (2012).
Gerald, D. et al. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell 118, 781–794 (2004).
Metzen, E., Zhou, J., Jelkmann, W., Fandrey, J. & Brune, B. Nitric oxide impairs normoxic degradation of HIF-1alpha by inhibition of prolyl hydroxylases. Mol. Biol. Cell 14, 3470–3481 (2003).
Briggs, K. J. et al. Paracrine induction of HIF by glutamate in breast cancer: EglN1 senses cysteine. Cell 166, 126–139 (2016).
Oh, E. T. et al. NQO1 inhibits proteasome-mediated degradation of HIF-1alpha. Nat. Commun. 7, 13593 (2016).
Allavena, P., Sica, A., Solinas, G., Porta, C. & Mantovani, A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit. Rev. Oncol. Hematol. 66, 1–9 (2008).
Chen, J. et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 19, 541–555 (2011).
Solinas, G. et al. Tumor-conditioned macrophages secrete migration-stimulating factor: a new marker for M2-polarization, influencing tumor cell motility. J. Immunol. 185, 642–652 (2010).
Ruffell, B. & Coussens, L. M. Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472 (2015).
Su, S. et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell 25, 605–620 (2014).
Semenza, G. L. Hypoxia-inducible factors: coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 36, 252–259 (2017).
Zhou, W. et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25, 501–515 (2014).
Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 (2015).
Liu, B. et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381 (2015).
Ostrowski, M. et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 (2010). sup pp 11-13.
Beatson, R. et al. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat. Immunol. 17, 1273–1281 (2016).
Lee, D. C. et al. A lactate-induced response to hypoxia. Cell 161, 595–609 (2015).
Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014).
Wang, W. et al. Effector T cells abrogate stroma-mediated chemoresistance in ovarian cancer. Cell 165, 1092–1105 (2016).
Lin, S. et al. Lactate-activated macrophages induced aerobic glycolysis and epithelial–mesenchymal transition in breast cancer by regulation of CCL5–CCR5 axis: a positive metabolic feedback loop. Oncotarget 8, 110426–110443 (2017).
Chen, P. et al. Gpr132 sensing of lactate mediates tumor–macrophage interplay to promote breast cancer metastasis. Proc. Natl Acad. Sci. USA 114, 580–585 (2017).
Goswami, S. et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 65, 5278–5283 (2005).
Hernandez, L. et al. The EGF/CSF-1 paracrine invasion loop can be triggered by heregulin beta1 and CXCL12. Cancer Res. 69, 3221–3227 (2009).
Yang, M. et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 10, 117 (2011).
Luga, V. et al. Exosomes mediate stromal mobilization of autocrine Wnt–PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 (2012).
Boelens, M. C. et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 159, 499–513 (2014).
Hoshino, A. et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015).
Costa-Silva, B. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015).
Huang, X. et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14, 319 (2013).
Melo, S. A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014).
Hay, N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Cancer 16, 635–649 (2016).
Lieberman, J. Manipulating the in vivo immune response by targeted gene knockdown. Curr. Opin. Immunol. 35, 63–72 (2015).
Wheeler, L. A. et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer–siRNA chimeras. J. Clin. Invest. 121, 2401–2412 (2011).
Song, E. et al. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 77, 7174–7181 (2003).
Liu, H. et al. Cysteine-rich protein 61 and connective tissue growth factor induce deadhesion and anoikis of retinal pericytes. Endocrinology 149, 1666–1677 (2008).
Sisto, M. et al. Fibulin-6 expression and anoikis in human salivary gland epithelial cells: implications in Sjogren’s syndrome. Int. Immunol. 21, 303–311 (2009).
Lee, H. W. et al. Tpl2 kinase impacts tumor growth and metastasis of clear cell renal cell carcinoma. Mol. Cancer Res. 11, 1375–1386 (2013).
Liu, Y. et al. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 30, 243–256 (2016).
Qu, L. et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016).
Ries, C. H. et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25, 846–859 (2014).
Acknowledgements
This work was supported by grants from the Natural Science Foundation of China (81490750), the National Key Research and Development Program of China (2016YFC1302300 and 2017YFA0106300), the Natural Science Foundation of China (81720108029, 81621004, 81622036 81472468, 81672614 and 81802645), the Guangdong Science and Technology Department (2016B030229004 and 2017B030314026), the Science Foundation of Guangdong Province (2016A030306023, 2017A030313878 and 201710010083), the Guangzhou Science Technology and Innovation Commission (201803040015), the Tip-top Scientific and Technical Innovative Youth Talents of Guangdong Special Support Program (no. 2016TQ03R553), and the Guangzhou Science Technology and Innovation Commission (201508020008 and 201508020249). This research is partly supported by the Fountain-Valley Life Sciences Fund of the University of Chinese Academy of Sciences Education Foundation.
Author information
Authors and Affiliations
Contributions
F.C., S.S., and E.S. conceived the ideas and designed the experiments. F.C., J.C., L.Y., J.L., X.Z., Y.Z., Q.T., D.H.,Y.X., and J.Z. performed the experiments. Q.L., F.S., and E.S. provided the patient samples for clinical data analysis.F.C., J.C., D.Y., D.L., P.-P.W., and M.L. analyzed the data. F.C. and E.S. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 TAMs enhance aerobic glycolysis and inhibit apoptosis of breast cancer cells, related to Figure 1.
a. Correlation of GLUT3 and CD68 expression in different subtypes of breast cancer samples. ER+HER2- group, n = 209; ER+HER2+ group, n = 103; ER-HER2+ group, n = 71; ER-HER2- group, n = 70. Spearman order correlation analysis was performed to assess the relationship. b, c. Correlation of the expression between GLUT1 and CD68 (b) or HK2 and CD68 (c) in breast cancer samples, n = 453. Spearman order correlation analysis was performed to assess the relationship. d. Quantification of TUNEL+ cells in breast cancer samples with high or low GLUT3 expression. GLUT3 low group, n = 108; GLUT3 high group, n = 49. Mean ± SEM and p value were determined by independent-sample t tests. e-q. Breast cancer cells were cocultured with MDMs or pol-TAMs in transwell systems for 6 days and then harvested for indicated experiments. n = 3 independent experiments. (e, g-q) Means ± SD and p value were determined by independent-sample t tests. e. ECAR of the MDA-MB-231 cells with indicated treatments. f. Western blotting for the expression of GLUT3, HK2, PKM2 and LDHA in MDA-MB-231 cells with indicated treatments. g-j. Glucose consumption (left) and lactate production (right) of the MDA-MB-231 cells (g), MDA-MB-468 cells (h), MCF-7 cells (i) and BT-474 cells (j) with indicated treatments. k-n. Quantification for the proportion of Annexin V+ apoptotic cells in the MDA-MB-231 cells treated with 30 mM Cisplatin for 24 h (k) or MDA-MB-468 cells (l), MCF-7 cells (m) and BT-474 cells (n) treated with 2 μg/ml docetaxel for 12 h. Annexin V/PI staining was determined by flow cytometry. o-q. Quantification for the proportion of cell death in the MDA-MB-468 cells (o), MCF-7 cells (p) and BT-474 cells (q) upon withdrawing extracellular matrix support. Calcein AM was used to detect cell viability and EthD-1 for cell death.
Supplementary Figure 2 TAMs enhance aerobic glycolysis and inhibit apoptosis of breast cancer cells by stabilizing HIF-1α protein, related to Figure 2.
a. Western blotting for the expression of HIF-1α in the MDA-MB-231 cells cocultured with MDMs or pol-TAMs, n = 3 independent experiments. b. Quantification for the expression kinetics of HIF-1α in the MDA-MB-231 cells treated with pol-TAM condition media, determined by Image J, related to Fig. 2a. c. mRNA expression of HIF-1α in the MDA-MB-231 cells cocultured with MDMs or pol-TAMs was determined by qRT-PCR. d-p. HIF-1α siRNAs were transfected into the breast cancer cells, which were cocultured with pol-TAMs in transwell systems for 6 days. Subsequently, breast cancer cells were harvested for indicated experiments. d-f. Glucose consumption (left) and lactate production (right) of the MDA-MB-468 cells (d), MCF-7 cells (e) and BT-474 cells (f) with indicated treatments. g-k. Quantification for the proportion of Annexin V+ apoptotic cells in the MDA-MB-231 cells treated with 2 μg/ml docetaxel for 12 h (g) or 30 mM Cisplatin for 24 h (h), MDA-MB-468 cells (i), MCF-7 cells (j) and BT-474 cells (k) treated with 2 μg/ml docetaxel for 12 h. Annexin V/PI staining was determined by flow cytometry. l-o. Quantification for the proportion of cell death in the MDA-MB-231 cells (l), MDA-MB-468 cells (m), MCF-7 cells (n) and BT-474 cells (o) by withdrawing extracellular matrix support. Calcein AM was used to detect cell viability and EthD-1 for cell death. p. Flow cytometric analysis for CellROX green in the MDA-MB-231 cells treated with 2 μg/ml docetaxel for 12hr. Mean fluorescence intensity (MFI) of CellROX green was shown. (c-p) n = 3 independent experiments, Mean ± SD and p value were determined by independent-sample t tests.
Supplementary Figure 3 TAMs enhance aerobic glycolysis of breast cancer cells by extracellular vesicle-packaged lncRNA, related to Figure 3.
a. The schematics of the approach for the EV purification. b. Glucose consumption (left) and lactate production (right) of the MDA-MB-231 cells treated with pol-TAM condition media (CM) or supernatant (S) and pellet (P) extracted from pol-TAM CM. c. Western blotting for Alix expression in EVs extracted by OptiprepTM density gradient separation. d. Particle diameter of EVs extracted from pol-TAM CM was determined by Nano sight. e. Western blotting for the expression of Alix, CD63 and CD81 in the pol-TAMs or EVs and supernatant extracted from the pol-TAM CM. f-r. Breast cancer cells treated with EVs or whole CM collected from pol-TAMs for 6 days were harvested for indicated experiments. Neutralizing antibody against CD81 was used to deplete EVs in the CM. f. Western blotting for the expression of HIF-1α, GLUT3, HK2, PKM2 and LDH-A in the MDA-MB-231 cells. g-i. Glucose consumption (left) and lactate production (right) of the MDA-MB-468 cells (g), MCF-7 cells (h) and BT-474 cells (i). j-n. Quantification for the proportion of Annexin V+ apoptotic cells in the MDA-MB-231 cells treated with 2 μg/ml docetaxel for 12 h (j) or 30 mM cisplatin for 24 h (k), MDA-MB-468 cells (l), MCF-7 cells (m) and BT-474 cells (n) treated with 2 μg/ml docetaxel for 12 h. Annexin V/PI staining was determined by flow cytometry. o-r. Quantification for the proportion of cell death in the MDA-MB-231 cells (o), MDA-MB-468 cells (p), MCF-7 cells (q) and BT-474 cells (r) by withdrawing extracellular matrix support. Calcein AM was used to detect cell viability and EthD-1 for cell death. s. RNA content in pol-TAMs and TAM EVs was detected by RNA sequencing. t. Particle diameter of EVs produced by pol-TAMs transduced with lenti-Rab27-shRNAs was determined by Nano sight. (b, c, e-r) n = 3 independent experiments. (b, g-r) Mean ± SD and p value were determined by independent-sample t tests.
Supplementary Figure 4 HISLA transmitted by TAM EVs enhances aerobic glycolysis and induces apoptosis resistance in breast cancer cells, related to Figure 4.
(a-d) Pol-TAMs were transduced with lenti-shRNAs against indicated lncRNAs and cocultured with MDA-MB-231 cells. a, b. Expression of indicated lncRNAs in the pol-TAMs EVs (a) or MDA-MB-231 cells (b) was determined by qRT-PCR. c, d. Glucose consumption (c) and lactate production (d) of the MDA-MB-231 cells. e. Western blotting for the expression of GLUT3, HK2, PKM2, LDHA and HIF-1α in the MDA-MB-231 cells. f. Glucose consumption (left) and lactate production (right) of the MDA-MB-231 cells cocultured with pol-TAMs transduced with lenti-RAB27-shRNAs. g. Western blotting for the expression of GLUT3, HK2, PKM2, LDHA and HIF-1α in the MDA-MB-231 cells as in f. h. Glucose consumption (left) and lactate production (right) of the MDA-MB-231 cells transfected with HISLA siRNAs and cocultured with pol-TAMs. i. Western blotting for the expression of GLUT3, HK2, PKM2, LDHA and HIF-1α in the MDA-MB-231 cells as in h. (j-l) MDA-MB-231 cells were ectopic expressed with HISLA. j. Expression of HISLA in the MDA-MB-231 cells was determined by qRT-PCR. k. Glucose consumption (left) and lactate production (right) of the MDA-MB-231 cells. l. Western blotting for the expression of GLUT3, HK2, PKM2, LDHA and HIF-1α in the MDA-MB-231 cells. m. Expression of HISLA in the MDA-MB-231 cells treated with pol-TAM EVs was determined by qRT-PCR. α–amanitin(10 μM) was used to inhibit the RNA transcription. n-o. Quantification of the migration (n) and invasion (o) of the MDA-MB-231 cells treated with EVs extracted from pol-TAMs transduced with lenti-HISLA-shRNAs. p. Expression of HISLA in EVs (left) and MDA-MB-231 cells treated with pol-TAM EVs (right) was determined by qRT-PCR. RNase(2 μg/ml) was used to degrade RNA and triton(0.1%) was used for membrane permeabilization in EVs. q. Northern blotting for the expression of HISLA in MCF-7, BT474, T47D, MDA-MB-231, BT549 and MDA-MB-436 cells treated without or with pol-TAMs EVs. (a-q) n = 3 independent experiments. (a-d, f, h, j, k, m-p) Mean ± SD and p value were determined by independent-sample t tests.
Supplementary Figure 5 Lactate in the tumor microenvironment upregulates HISLA in TAMs, related to Figure 5.
a. Northern blotting for the expression of HISLA in indicated cell types (n = 3 independent experiments). b. Luciferase reporter assays for THP-1 cells with endogenous PU.1 expression transfected with reporter plasmids containing serious deletion HISLA promoter constructs. c. Schematic graphic for online dataset of PU.1 ChIP-seq. d. Expression of HISLA in MDMs treated with MDA-MB-231 cell CM or lactate was determined by qRT-PCR. e. Glucose consumption (left) and lactate production (right) of MDMs or pol-TAMs transduced with lenti-HISLA-shRNAs and treated with CM extracted from MDA-MB-231 cells. f. Glucose consumption (left) and lactate production (right) of MDMs or pol-TAMs with indicated treatments. Neutralizing antibodies against GM-CSF or CD81 were used to offset GM-CSF effect or to deplete EVs. Oxamate at 90 mM or DCA at 2 mM was used to inhibit lactate secretion from the MDA-MB-231 cells before collecting the CM. g. HEK293T cells were co-transfected with PU.1 expression plasmid and reporter plasmids containing PU.1-regulatory site. The cells were treated with lactate and luciferase reporter assays was performed. h. Luciferase reporter assays for the pol-TAMs transfected with reporter plasmids containing PU.1-regulatory site and co-cultured with MDA-MB-231 cells. i. A conserved ELK1-binding element on the promoter of HISLA was predicted by JASPAR. j. Western blotting for the phosphorylation of ERK in the pol-TAMs with indicated treatments. k. Expression of HISLA in the pol-TAMs treated with ERK inhibitors(SCH772984:1 μM, FR180204: 10 μM) and CM extracted from MDA-MB-231 cells was determined by qRT-PCR. (a, b, d-h, j, k) n = 3 independent experiments. (b, d-h, k) Mean ± SD and p value were determined by independent-sample t tests.
Supplementary Figure 6 HISLA stabilizes HIF-1α by interfering PHD2 and HIF-1α interaction, related to Figure 6.
a-c. MDA-MB-231 cells were transfected with HISLA-siRNAs and then cocultured with pol-TAMs. HIF-1α ubiquitination in the cocultured MDA-MB-231 cells was determined by immunoprecipitation (a). The hydroxylation of HIF-1α and the expression of PHD2 and VHL in the MDA-MB-231 cells was determined by western blotting (b). The interaction between PHD2 and HIF-1α in the MDA-MB-231 cells was determined by immunoprecipitation (c). MG132(20 μM) was added prior to protein extraction to inhibit HIF-1α degradation. d-h. MDA-MB-231 cells were ectopic expressed with HISLA. d. The interaction between PHD2 and HIF-1α in the MDA-MB-231 cells was determined by immunoprecipitation. e. The hydroxylation of HIF-1α and the expression of PHD2 and VHL in the MDA-MB-231 cells was determined by western blotting. MG132(20 μM) was added prior to protein extraction to inhibit HIF-1α degradation. f. HIF-1α ubiquitination in the MDA-MB-231 cells was determined by immunoprecipitation. MG132(20 μM) was added prior to protein extraction to inhibit HIF-1α degradation. g. The interaction between HISLA and PHD2, HIF-1α or VHL in the MDA-MB-231 cells was determined by RNA pull down. h. The binding of HISLA with PHD2, HIF-1α or VHL in the MDA-MB-231 cells cocultured with pol-TAMs was determined by RNA immunoprecipitation. i. HISLA expression in the nuclear and cytoplasmic extracts of MDA-MB-231 cells treated with pol-TAM EVs was determined by qRT-PCR. j, k. The in vitro interaction between HISLA and HIF-1α (j) or PHD2 (k) was determined by RNA pull down. l. Prediction of the secondary structure of HISLA and HISLA190–248 by Mfold software. m. The interaction between HISLA truncations and PHD2 was determined by RNA pull down. (a-k,m) n = 3 independent experiments. (h,i) Mean ± SD was determined by independent-sample t tests.
Supplementary Figure 7 TAMs release HISLA in extracellular vesicles to enhance glycolysis and chemoresistance of breast cancer in vivo, related to Figure 7.
a, b. Quantification of SUV for the glucose consumption of the xenografts with indicated treatments. (a) related to Fig.7b, (b) related to Fig.7f. n = 5 mice per group. c, d. mRNA expression of GLUT1, GLUT3 and HK2 in the xenografts with indicated treatments was determined by qRT-PCR. n = 5 xenografts per group in c and n = 3 xenografts per group in d. e, f. Western blotting for HIF-1α in the xenografts with indicated treatments. n = 5 xenografts per group in e and n = 3 xenografts per group in f. g, h. Representative H&E staining for the lung tissue sections obtained from mice with indicated treatments. (g) related to Fig.7d, (h) related to Fig.7h. Scale bars, 50 μm. n = 5 mice per group. i, j. Quantification for the photon flux of lung metastasis in the mice with indicated treatments. (i) related to Fig.7d, (j) related to Fig.7h. Mean ± SD, n = 5 mice per group. (a-d, i, j) Mean ± SD and p value were determined by independent-sample t tests.
Supplementary Figure 8 HISLA correlates with tumor glycolysis and chemoresistance of breast cancer patients, related to Figure 8.
a. Correlation between the expression of HISLA and CD68 in breast cancer samples. HISLA was detected by ISH staining and CD68 was detected by IHC staining. Spearman order correlation analysis was performed, n = 453, r = 0.577 and p < 0.0001. b. Kaplan-Meier survival curves for breast cancer patients with high/low GLUT3 expression in different breast cancer subtypes.
Supplementary Figure 9 Uncropped blots for western blotting and northern blotting.
Most uncropped blot were detected by GBOX EXTENDED Chemi XT 4, others were detected by western blot film.
Supplementary information
Supplementary Information
Supplementary Figures 1–9 and legends for Supplementary Tables 1–6.
Supplementary Table 1
Correlation of glycolytic enzymes and CD68 expression in Breast Cancer in the Oncomine Online Database
Supplementary Table 2
Differential mRNA expression in the lncRNA microarray as in Fig. 3e.
Supplementary Table 3
Correlation between the expression of HISLA and indicated glycolytic enzymes in breast cancer samples.
Supplementary Table 4
Correlation of HISLA expression with clinicopathological status in 453 cases.
Supplementary Table 5
Sequences for si/shRNAs, LNA probe, primer sequences for qRT-PCR and ChIP assay.
Supplementary Table 6
Statistics source data.
Rights and permissions
About this article
Cite this article
Chen, F., Chen, J., Yang, L. et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol 21, 498–510 (2019). https://doi.org/10.1038/s41556-019-0299-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0299-0
This article is cited by
-
The roles and molecular mechanisms of non-coding RNA in cancer metabolic reprogramming
Cancer Cell International (2024)
-
Novel lncRNA Gm33149 modulates metastatic heterogeneity in melanoma by regulating the miR-5623-3p/Wnt axis via exosomal transfer
Cancer Gene Therapy (2024)
-
Tumour-associated macrophages and Schwann cells promote perineural invasion via paracrine loop in pancreatic ductal adenocarcinoma
British Journal of Cancer (2024)
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
Nanoscale metal–organic frameworks as smart nanocarriers for cancer therapy
Journal of Nanostructure in Chemistry (2024)