Abstract
Recent studies have revealed a role for macrophages and neutrophils in limiting chemotherapy efficacy; however, the mechanisms underlying the therapeutic benefit of myeloid-targeting agents in combination with chemotherapy are incompletely understood. Here, we show that targeting tumour-associated macrophages by colony-stimulating factor-1 receptor (CSF-1R) blockade in the K14cre;Cdh1F/F;Trp53F/F transgenic mouse model for breast cancer stimulates intratumoural type I interferon (IFN) signalling, which enhances the anticancer efficacy of platinum-based chemotherapeutics. Notably, anti-CSF-1R treatment also increased intratumoural expression of type I IFN-stimulated genes in patients with cancer, confirming that CSF-1R blockade is a powerful strategy to trigger an intratumoural type I IFN response. By inducing an inflamed, type I IFN-enriched tumour microenvironment and by further targeting immunosuppressive neutrophils during cisplatin therapy, antitumour immunity was activated in this poorly immunogenic breast cancer mouse model. These data illustrate the importance of breaching multiple layers of immunosuppression during cytotoxic therapy to successfully engage antitumour immunity in breast cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The RNA-seq data derived from mouse samples that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE101881. Source data for Figs. 1a,d–i, 2, 3c–h, 4, 5 and 6 and Supplementary Figs. 1a,b,e,f,h,i, 2, 3e–h, 5a–c,f–i and 6a–f,i–n have been provided as Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Coffelt, S. B. & de Visser, K. E. Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends Immunol. 36, 198–216 (2015).
Galluzzi, L., Buque, A., Kepp, O., Zitvogel, L. & Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 17, 97–111 (2017).
Ciampricotti, M., Hau, C. S., Doornebal, C. W., Jonkers, J. & de Visser, K. E. Chemotherapy response of spontaneous mammary tumors is independent of the adaptive immune system. Nat. Med. 18, 344–346 (2012).
DeNardo, D. G. et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1, 54–67 (2011).
Ruffell, B. et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 26, 623–637 (2014).
Campbell, M. J. et al. The prognostic implications of macrophages expressing proliferating cell nuclear antigen in breast cancer depend on immune context. PLoS One 8, e79114 (2013).
Chua, W., Charles, K. A., Baracos, V. E. & Clarke, S. J. Neutrophil/lymphocyte ratio predicts chemotherapy outcomes in patients with advanced colorectal cancer. Br. J. Cancer 104, 1288–1295 (2011).
Kishi, Y. et al. Blood neutrophil-to-lymphocyte ratio predicts survival in patients with colorectal liver metastases treated with systemic chemotherapy. Ann. Surg. Oncol. 16, 614–622 (2009).
Miao, Y., Yan, Q., Li, S., Li, B. & Feng, Y. Neutrophil to lymphocyte ratio and platelet to lymphocyte ratio are predictive of chemotherapeutic response and prognosis in epithelial ovarian cancer patients treated with platinum-based chemotherapy. Cancer Biomark. 17, 33–40 (2016).
Pistelli, M. et al. Pre-treatment neutrophil to lymphocyte ratio may be a useful tool in predicting survival in early triple negative breast cancer patients. BMC Cancer 15, 195 (2015).
Paulus, P., Stanley, E. R., Schafer, R., Abraham, D. & Aharinejad, S. Colony-stimulating factor-1 antibody reverses chemoresistance in human MCF-7 breast cancer xenografts. Cancer Res. 66, 4349–4356 (2006).
Shree, T. et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 25, 2465–2479 (2011).
Mitchem, J. B. et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 73, 1128–1141 (2013).
Di Mitri, D. et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature 515, 134–137 (2014).
Houthuijzen, J. M. et al. Lysophospholipids secreted by splenic macrophages induce chemotherapy resistance via interference with the DNA damage response. Nat. Commun. 5, 5275 (2014).
Acharyya, S. et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150, 165–178 (2012).
Olson, O. C., Kim, H., Quail, D. F., Foley, E. A. & Joyce, J. A. Tumor-associated macrophages suppress the cytotoxic activity of antimitotic agents. Cell Rep. 19, 101–113 (2017).
Weizman, N. et al. Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene 33, 3812–3819 (2014).
Nakasone, E. S. et al. Imaging tumor–stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell 21, 488–503 (2012).
Nywening, T. M. et al. Targeting both tumour-associated CXCR2+ neutrophils and CCR2+ macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut 67, 1112–1123 (2018).
Ries, C. H. et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25, 846–859 (2014).
Quail, D. F. & Joyce, J. A. Molecular pathways: deciphering mechanisms of resistance to macrophage-targeted therapies. Clin. Cancer Res. 23, 876–884 (2017).
Crusz, S. M. & Balkwill, F. R. Inflammation and cancer: advances and new agents. Nat. Rev. Clin. Oncol. 12, 584–596 (2015).
Derksen, P. W. et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10, 437–449 (2006).
Hume, D. A. & MacDonald, K. P. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood 119, 1810–1820 (2012).
Franklin, R. A. et al. The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 (2014).
Doornebal, C. W. et al. A preclinical mouse model of invasive lobular breast cancer metastasis. Cancer Res. 73, 353–363 (2013).
Noy, R. & Pollard, J. W. Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61 (2014).
Bradley, E. W., Ruan, M. M. & Oursler, M. J. Novel pro-survival functions of the Kruppel-like transcription factor Egr2 in promotion of macrophage colony-stimulating factor-mediated osteoclast survival downstream of the MEK/ERK pathway. J. Biol. Chem. 283, 8055–8064 (2008).
Gomez-Roca, C. A. et al. Phase I study of RG7155, a novel anti-CSF1R antibody, in patients with advanced/metastatic solid tumors. J. Clin. Oncol. 33, 3005 (2015).
Zitvogel, L., Galluzzi, L., Kepp, O., Smyth, M. J. & Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 (2015).
Vacchelli, E. et al. Autocrine signaling of type 1 interferons in successful anticancer chemotherapy. Oncoimmunology 4, e988042 (2015).
Parker, B. S., Rautela, J. & Hertzog, P. J. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer 16, 131–144 (2016).
Snell, L. M., McGaha, T. L. & Brooks, D. G. Type I interferon in chronic virus infection and cancer. Trends Immunol. 38, 542–557 (2017).
Fuertes, M. B. et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells. J. Exp. Med. 208, 2005–2016 (2011).
Coffelt, S. B. et al. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 (2015).
Sistigu, A. et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 20, 1301–1309 (2014).
Burnette, B. C. et al. The efficacy of radiotherapy relies upon induction of type I interferon-dependent innate and adaptive immunity. Cancer Res. 71, 2488–2496 (2011).
Deng, L. et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014).
Bidwell, B. N. et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat. Med. 18, 1224–1231 (2012).
Callari, M. et al. Subtype-dependent prognostic relevance of an interferon-induced pathway metagene in node-negative breast cancer. Mol. Oncol. 8, 1278–1289 (2014).
Snijders, A. M. et al. An interferon signature identified by RNA-sequencing of mammary tissues varies across the estrous cycle and is predictive of metastasis-free survival. Oncotarget 5, 4011–4025 (2014).
Schiavoni, G. et al. Cyclophosphamide synergizes with type I interferons through systemic dendritic cell reactivation and induction of immunogenic tumor apoptosis. Cancer Res. 71, 768–778 (2011).
Weichselbaum, R. R. et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl Acad. Sci. USA 105, 18490–18495 (2008).
Critchley-Thorne, R. J. et al. Impaired interferon signaling is a common immune defect in human cancer. Proc. Natl Acad. Sci. USA 106, 9010–9015 (2009).
Pyonteck, S. M. et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 (2013).
Quail, D. F. et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 352, aad3018 (2016).
Yan, D. et al. Inhibition of colony stimulating factor-1 receptor abrogates microenvironment-mediated therapeutic resistance in gliomas. Oncogene 36, 6049–6058 (2017).
Zhu, Y. et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 74, 5057–5069 (2014).
Coffelt, S. B., Wellenstein, M. D. & de Visser, K. E. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446 (2016).
Pylaeva, E., Lang, S. & Jablonska, J. The essential role of type I interferons in differentiation and activation of tumor-associated neutrophils. Front. Immunol. 7, 629 (2016).
Berry, M. P. et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 (2010).
Rocha, B. C. et al. Type I interferon transcriptional signature in neutrophils and low-density granulocytes are associated with tissue damage in malaria. Cell Rep. 13, 2829–2841 (2015).
Teijaro, J. R. et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211 (2013).
Cunningham, C. R. et al. Type I and type II interferon coordinately regulate suppressive dendritic cell fate and function during viral persistence. PLoS Pathog. 12, e1005356 (2016).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Michaut, M. et al. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci. Rep. 6, 18517 (2016).
Schouten, P. C. et al. Robust BRCA1-like classification of copy number profiles of samples repeated across different datasets and platforms. Mol. Oncol. 9, 1274–1286 (2015).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Salvagno, C. & de Visser, K. E. Purification of immune cell populations from freshly isolated murine tumors and organs by consecutive magnetic cell sorting and multi-parameter flow cytometry-based sorting. Methods Mol. Biol. 1458, 125–135 (2016).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Oesper, L., Merico, D., Isserlin, R. & Bader, G. D. WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol. Med. 6, 7 (2011).
Spinelli, L., Carpentier, S., Montanana Sanchis, F., Dalod, M. & Vu Manh, T. P. BubbleGUM: automatic extraction of phenotype molecular signatures and comprehensive visualization of multiple gene set enrichment analyses. BMC Genomics 16, 814 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Pradel, L. P. et al. Macrophage susceptibility to emactuzumab (RG7155) treatment. Mol. Cancer Ther. 15, 3077–3086 (2016).
Acknowledgements
This work was supported by the European Union (FP7 MCA-ITN 317445 TIMCC), the Dutch Cancer Society (NKI10623), the European Research Council (ERC consolidator award INFLAMET 615300), Worldwide Cancer Research (AICR 11-0677), the Netherlands Organization for Scientific Research NWO VIDI (917.96.307) and Oncode. K. Kos is supported by an OOA/NWO Diamond grant. K.E.d.V. is an EMBO Young Investigator. J.L.S. is a member of the Excellence Cluster ImmunoSensation and is in part supported by the DFG (SFB704, Excellence Cluster ImmunoSensation). We thank M. D. Wellenstein, H. Garner, S. Bissinger, J. Borst and T. Schumacher for useful discussions. We thank M. Hauptmann for advice on statistical analyses on the mouse survival curves. We thank the clinical investigators J.-Y. Blay, C. Gomez-Roca, J.-P. Delord, M. Toulmond, C. le Tourneau and A. Italiano for running clinical trials with emactuzumab, and M. Cannarile, B. Quackenbush and A. Jegg for translational medicine support at Roche. We thank the histology core facility within the Cancer Research UK Beatson Institute for performing NKp46 immunohistochemistry on mouse tumour tissue. We thank K. Wartha, S. Klarenbeek and I. Peters Rit for technical assistance and the researchers involved in the RATHER project for generously providing tissue sections of human ILCs. We thank the flow cytometry facility, the animal facility, the animal pathology facility and the Core Facility Molecular Pathology and Biobanking at the NKI.
Author information
Authors and Affiliations
Contributions
C.S., M.C., C.H.R., J.J. and K.E.d.V. conceived the ideas and designed the experiments. C.S., M.C., C.-S.H., S.B.C., K. Kersten, A.v.W., K.V., K. Kos and K.E.d.V. performed the experiments and analysed the data. C.H.R. provided the anti-CSF-1R antibody and control antibody. S.T., T.U. and J.L.S. prepared the samples and conducted the RNA-seq and bioinformatics analyses on murine cells. J.-Y.S. performed the metastasis scoring. C.-H.O., D.R. and P.A.C. were involved in the collection of patient samples and bioinformatic analysis of the human data. C.S. and K.E.d.V. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
C.H.R., C.-H.O. and D.R. are employees of F. Hoffman La Roche. C.H.R. is an inventor of granted and pending patent applications relating to emactuzumab and a stockholder in F. Hoffman La Roche. F. Hoffman La Roche provided financial research support for the experiments with anti-CSF-1R. P.A.C. received funding from Roche for the described clinical trial and other Roche-sponsored studies.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
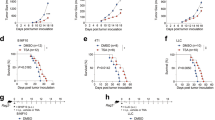
Supplementary Figure 1 CSF-1R blockade does not influence spontaneous metastasis formation.
(a) Presence of CD68+ macrophages in untreated human invasive lobular carcinomas and in adjacent normal breast tissue. Influx of CD68+ macrophages was scored based on immunohistochemistry (normal breast tissue, n=5 patients; invasive lobular carcinomas, n=14 patients). Representative images are shown. Scale bar=25μm. (b) Representative IHC staining of F4/80+ macrophages in a mammary tumor of a KEP mouse and in a normal mammary gland of an age-matched WT mouse. Scale bar=20μm. Percentage of CD11b+F4/80+ macrophages gated on CD45+ cells in KEP mammary tumors (n=20 animals) and in normal mammary glands of age-matched WT mice (n=4 animals). (c) Representative flow cytometry histogram and plot showing CD206 and MHCII expression, respectively, on F4/80+ macrophages in a KEP mammary tumor. Data are representative of 5 independent KEP mammary tumors. (d) Flow cytometry histograms showing CSF-1R expression levels (white) and Fluorescence minus one (FMO) control (grey) in 3 independent KEP mammary tumors. (e) Proportion of CD45+ immune cells of total live cells in tumors of time point-sacrificed KEP mice treated with control ab (n=6 animals) or anti-CSF-1R (n=5 animals) as determined by flow cytometry. (f) Organs collected from KEP mice bearing end-stage mammary tumors treated with control ab (n=20 animals) or anti-CSF-1R (n=22 animals) were microscopically analyzed for the presence of metastases. Percentage of tumor-bearing KEP mice with metastases is displayed. p=0.1 by Fisher’s exact test (Two-sided). (g) Schematic overview of continuous and adjuvant antibody treatment in the KEP-based spontaneous metastasis model as described in Methods. (h) Kaplan-Meier metastasis-specific survival curves of recipient mice orthotopically transplanted with tumor fragments from KEP mice and treated either continuously (control ab n=13 animals, anti-CSF-1R n=14 animals) or in an adjuvant setting (control ab n=13 animals, anti-CSF-1R n=11 animals). An event is defined as an animal that was sacrificed because of clinical signs of metastatic disease. (i) Quantification of the number of spontaneous pulmonary metastases in mice treated either continuously (control ab n=9 animals; anti-CSF-1R n=8 animals) or in an adjuvant setting (Control ab n=12 animals; anti-CSF-1R n=9 animals). Data presented in b, e and i are mean ± SEM and statistical analysis was performed using two-tailed Mann–Whitney test.
Supplementary Figure 2 CSF-1R blockade synergizes with platinum-based chemotherapy drugs, and not with docetaxel.
(a-b) Proportion of CD45+ immune cells gated on live cells (a) and F4/80+Ly6G- macrophages gated on CD11b+ cells (b) determined by flow cytometry in tumors of time point-sacrificed KEP mice treated as indicated (n=5 animals/group). (c) Kaplan Meier tumor-specific survival curves of KEP mice treated with control ab, anti-CSF-1R (same groups as Fig. 1f), docetaxel/control ab (n=10 animals) or docetaxel/anti-CSF-1R (n=10 animals). Docetaxel/control ab versus Control ab, p=0.0021; Docetaxel/control ab versus docetaxel/anti-CSF-1R, p=0.329 (two-tailed log-rank test). (d) Quantification of cleaved caspase 3+ cells in viable areas of mammary tumors of time point-sacrificed KEP mice treated with control ab (n=6 animals), anti-CSF-1R (n=5 animals), cisplatin/control ab (n=7 animals) and cisplatin/anti-CSF-1R (n=9 animals) as determined by IHC. (e) Quantification of CD31+ vessels in viable areas of mammary tumors of time point-sacrificed KEP mice treated with control ab (n=5 animals), anti-CSF-1R (n=5 animals), cisplatin/control ab (n=5 animals) and cisplatin/anti-CSF-1R (n=4 animals) as determined by immunofluorescence. Values represent average number of positive cells per FOV quantified by counting six fields per tumor. (f) Percentage of vessels covered by alpha-SMA+ pericytes in viable areas of mammary tumors as determined by immunofluorescence. Same mice as e. Percentage was determined by counting alpha-SMA+CD31+ cells and total CD31+ cells in six high-power microscopic fields per tumor. (g-h) Quantification of γH2AX+ cells (g) and cisplatin adducts+ cells (h) in viable areas of mammary tumors of time point-sacrificed KEP mice treated as indicated (γH2AX: control ab n=6 animals, anti-CSF-1R n=4 animals, cisplatin/control ab n=7 animals, cisplatin/anti-CSF-1R n=8 animals; CIS-adducts: cisplatin/control ab n=5 animals, cisplatin/anti-CSF-1R n=6 animals). (i) Percentage of non-viable area per tumor section of time point-sacrificed KEP mice quantified by digital area analysis of H&E stained sections (n=5 animals/group). (j-m) Quantification of CD34+ cells (j), cleaved caspase 3+ cells (k), BrdU+ cells (l), γH2AX+ cells (m) in viable areas of mammary tumors of time point-sacrificed KEP mice treated as indicated (CD34: docetaxel/control ab n=5 animals, docetaxel/anti-CSF-1R n=3 animals; cCasp3: n=4 animals/group; BrdU: n=5 animals/group; γH2AX: n=4 animals/group). (n) Proportion of CD11b+F4/80+ macrophages gated on CD45+ cells as determined by flow cytometry in tumors of end-stage KEP mice treated as indicated (oxaliplatin/Control ab treatment n=6 animals; oxaliplatin/anti-CSF-1R treatment n=5 animals). Data presented in d, g-h, j-m show average number of positive cells per field of view (FOV) quantified by counting five high-power microscopic fields per tumor. Data presented in a-b and d-n are mean values ± SEM and statistical analysis was performed using two-tailed Mann–Whitney test. DOCE, docetaxel, CIS, cisplatin, OX, oxaliplatin.
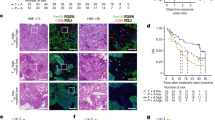
Supplementary Figure 3 Impact of CSF-1R inhibition on the intratumoral presence of diverse myeloid immune cell types.
(a) Representative dot plots of a KEP mammary tumor illustrating the gating strategy for the identification of cell populations. Antibody panel used: “tumor panel I” (see supplementary Table 1). Arrows indicate directionality of sub-gates. (b-d) tdTomato+ (Lineage-SiglecF-cKIT-CD11bintLy6G-Ly6C+) monocytes were isolated from the bone marrow of mTmG mice and adoptively transferred into tumor-bearing KEP mice that had previously received either control ab or anti-CSF-1R. 4 days after the monocyte transfer, the presence and phenotype of tdTomato+ cells in tumors were analyzed. (b) Gating strategy showing intratumoral tdTomato+ cells that express F4/80 in control ab- or anti-CSF-1R-treated recipient KEP mice. (c) Representative flow cytometry histograms showing CX3CR1, PD-L1, CCR2 and CD80 expression in tdTomato+ and tdTomato- macrophages in KEP tumors. (d) Overlay of representative dot plots showing Ly6C expression in tdTomato+ and tdTomato- macrophages in control ab- and anti-CSF-1R-treated KEP mice. Data presented in b-d are representative of 2 (control ab treatment) and 3 (anti-CSF-1R treatment) independent experiments. (e) Quantification of Ly6G+ neutrophils in viable areas of mammary tumors of time point-sacrificed KEP mice treated with control ab (n=6 animals), anti-CSF-1R (n=6 animals), cisplatin/control ab (n=7 animals) or cisplatin/anti-CSF-1R (n=7 animals). (f) Proportion of Ly6C+Ly6G- monocytes determined by flow cytometry in KEP mammary tumors treated with control ab (n=4 animals) or anti-CSF-1R (n=5 animals). (g) Quantification of Major Basic Protein (MBP)+ cells in viable areas of mammary tumors of time point-sacrificed KEP mice as determined by IHC (control ab n=5 animals, anti-CSF-1R n=6 animals, cisplatin/control ab n=7 animals, cisplatin/anti-CSF-1R n=7 animals) and proportion of Siglec F+ eosinophils gated on intratumoral CD45+ cells of time point-sacrificed KEP mice as determined by flow cytometry (cisplatin/control ab n=5 animals, cisplatin/anti-CSF-1R n=6 animals). (h) Quantification of Toluidine Blue+ mast cells in viable areas of mammary tumors of time point-sacrificed KEP mice (control ab n=4 animals, anti-CSF-1R n=6 animals, cisplatin/control ab n=7 animals, cisplatin/anti-CSF-1R n=7 animals) as determined by histochemistry. Values in e, g and h represent average number of positive cells per field of view (FOV) quantified by counting five high-power microscopic fields per tumor. Data presented in e-h are mean values ± SEM. Statistical analysis was performed using two-tailed Mann–Whitney test.
Supplementary Figure 4 Intratumoral neutrophils show elevated expression levels of type I IFN-stimulated genes upon CSF-1R blockade.
(a) Hierarchical clustering of the top 400 variable genes between neutrophils isolated from tumors of KEP mice treated with cisplatin/control ab (n=3 biologically independent samples) and cisplatin/anti-CSF-1R (n=4 biologically independent samples). Mice were sacrificed one day after second cisplatin injection. FC: ≥ 1,5; unadjusted p-value: ≤ 0,05. Statistical analysis was performed using two-way ANOVA. (b) Gene set enrichment analysis (GSEA) of Egr2 target genes obtained from RNA-Seq data in tumor-infiltrating neutrophils of anti-CSF-1R-treated KEP mice compared to control ab-treated KEP mice (n=4 animals/group). Enrichment scores were calculated using a weighted Kolmogorov–Smirnov-like statistic. (c) Network visualization of GOEA of the top 100 up-regulated and top 100 down-regulated genes (cisplatin/anti-CSF-1R vs. cisplatin/control ab neutrophils; FC: 1,5, unadjusted p-value: ≤ 0.05) using BiNGO and EnrichmentMap. Red and blue nodes represent the positively and negatively enriched GO-terms, respectively. Node size represents corresponding enrichment p-values (FDR corrected p-value: ≤ 0.05). The genes used as input for the BiNGO analysis are derived from the 2-way ANOVA model. The enrichment score was calculated with a hypergeometric statistical test, multiple testing correction was performed with the Benjamin & Hochberg FDR correction. (d) Top three canonical pathways identified using ingenuity pathway analysis (IPA) enriched in neutrophils isolated from cisplatin/anti-CSF-1R-treated tumors (n=4 biologically independent samples) compared to neutrophils from cisplatin/control ab-treated tumors (n=3 biologically independent samples). Statistical analysis was performed with standard IPA software statistics.
Supplementary Figure 5 CSF-1R blockade increases intratumoral Ifna expression.
(a) Transcripts of Ifna and Ifnb in KEP mammary tumors were determined by qPCR and normalized to β-actin (n=5 animals/group). Mice were analyzed one day after the second docetaxel injection. Graphs show the mean ± SEM in ΔCt values. (b-c) Transcripts of Isg15 and Oas1a in orthotopically transplanted K14cre;Trp53F/F (KP) tumors (Isg15: control ab n=7 animals, anti-CSF-1R n=8 animals; Oas1a: n=8 animals/group) (b) and subcutaneous MC38 tumors (n=8 animals/group) (c) treated as indicated were determined by qPCR and normalized to β-actin. Mice were analysed at a tumor size of 100mm2 (KP) or after 12 days from the start of the treatment (MC38). Graphs show the mean ± SEM in ΔCt values. (d) Representative dot plots of a KEP tumor illustrating the gating strategy for cell sorting by flow cytometry. After cell separation based on CD11b expression by magnetic columns, the CD11b+ and CD11b- fractions were stained as described in Methods followed by flow cytometry-based sorting of intratumoral cell populations. (e) Representative dot plots of a KEP tumor illustrating the gating strategy for the identification of pDCs. Antibody panel “tumor panel II” was used. Arrows indicate directionality of sub-gates. (f) Proportion of plasmacytoid dendritic cells (pDCs) in mammary tumors of end-stage KEP mice as determined by flow cytometry (control ab n=5 animals, anti-CSF-1R n=3 animals, cisplatin/control ab n=5 animals, cisplatin/anti-CSF-1R n=4 animals). (g-h) Transcripts of Ifna and Ifnb in CD11b+F4/80-Ly6G-Ly6C+ monocytes (Ifna and Ifnb: n=4 animals/group), CD45+CD11b-CD11c- lymphocytes (Ifna: n=4 animals/group; Ifnb: cisplatin/control ab n=3 animals, cisplatin/anti-CSF-1R n=4 animals), CD11b+F4/80-Ly6G+Ly6Clow neutrophils (Ifna and Ifnb: cisplatin/control ab n=2 animals, cisplatin/anti-CSF-1R n=4 animals) and CD45-CD11b-CD31- tumor cells/fibroblasts (Ifna: n=4 animals/group; Ifnb: cisplatin/control ab n=4 animals, cisplatin/anti-CSF-1R n=3 animals) isolated from end-stage KEP tumors were determined by qPCR and normalized to β-actin. (i) Transcript of Ifna in cultured bone marrow-derived macrophages treated for 24h with either control antibody or anti-CSF-1R in the presence of KEP cancer cell line-derived conditioned medium. Data are representative of 4 independent experiments. Data presented in a-c and f-h are mean values ± SEM and statistical analysis was performed using two-tailed Mann–Whitney test. CIS, cisplatin; DOCE, docetaxel.
Supplementary Figure 6 Neutrophil inhibition enhances intratumoral granzyme B expression and improves the synergistic anti-cancer effect of cisplatin/anti-CSF-1R in K14cre;Cdh1F/F;Trp53F/F mice.
(a-c) Quantification of CD8+ T cells (a), CD4+ T cells (b) and FoxP3+ regulatory T cells (c) in viable areas of mammary tumors of time point-sacrificed KEP mice (CD8: cisplatin/control ab n=7 animals, cisplatin/anti-CSF-1R n=7 animals, cisplatin/anti-CSF-1R/anti-Ly6G n=7 animals; CD4: cisplatin/control ab n=7 animals, cisplatin/anti-CSF-1R n=6 animals, cisplatin/anti-CSF-1R/anti-Ly6G n=6 animals; FoxP3: cisplatin/control ab n=7 animals, cisplatin/anti-CSF-1R n=7 animals, cisplatin/anti-CSF-1R/anti-Ly6G n=6 animals). (d) Proportion of granzyme B+ CD45+ lymphocytes (lymphocyte gate was based on SSC and FSC) determined by flow cytometry in the tumor of time point-sacrificed KEP mice treated as indicated (cisplatin/control ab n=3 animals, cisplatin/anti-CSF-1R n=4 animals, cisplatin/anti-CSF-1R/anti-Ly6G n=5 animals). (e-f) Quantification of S100A9+ cells in viable areas of mammary tumors (e) and lung (f) of end-stage KEP mice treated with cisplatin/anti-CSF-1R (n=8 animals) or cisplatin/anti-CSF-1R/anti-Ly6G (n=6 animals). (g-h) Representative tumor growth graphs of six individual KEP mice treated with cisplatin/anti-CSF-1R (g) and cisplatin/anti-CSF-1R/anti-Ly6G (h). Data are representative of 16 cisplatin/anti-CSF-1R-treated mice and 10 cisplatin/anti-CSF-1R/anti-Ly6G-treated mice. Red arrows indicate cisplatin injections. In pink, growth curve of a secondary tumor that developed in another mammary gland during the treatment. (i) Kaplan-Meier tumor-specific survival curves of KEP mice treated with control ab, anti-CSF-1R (same groups as Fig. 1f) or anti-CSF-1R/anti-Ly6G (n=10 animals). (j-n) Quantification of BrdU+ (j) γH2AX+ cells (k), cleaved caspase 3+ cells (l), CD31+ vessels (m) and cisplatin adducts+ cells (n) in viable areas of mammary tumors of time point-sacrificed KEP mice treated with cisplatin/anti-CSF-1R (same as Fig. 1h and Supplementary Fig. 2) and cisplatin/anti-CSF-1R/anti-Ly6G (BrdU n=6 animals; γH2AX n=5 animals; cCasp3 n=8 animals; CD31 n=5 animals; CIS adducts n=5 animals). Values presented in a-c, e-f, j-l and n represent average number of positive cells per field of view (FOV) as determined by IHC quantified by counting five high-power microscopic fields per tumor. Values presented in m represent average number of positive cells per FOV as determined by immunofluorescence by counting six field per tumor. Data presented in a-f and j-n are mean values ± SEM. Statistical analysis was performed using two-tailed Mann–Whitney test. CIS, cisplatin.
Supplementary Information
Supplementary Information
Supplementary Figures 1–6 and Supplementary Table 1–4 legends
Supplementary Table 1
List of antibodies.
Supplementary Table 2
List of primer sequences used for RT-PCR.
Supplementary Table 3
List of type I IFN-related pathways and genes.
Supplementary Table 4
Statistics source data.
Rights and permissions
About this article
Cite this article
Salvagno, C., Ciampricotti, M., Tuit, S. et al. Therapeutic targeting of macrophages enhances chemotherapy efficacy by unleashing type I interferon response. Nat Cell Biol 21, 511–521 (2019). https://doi.org/10.1038/s41556-019-0298-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0298-1
This article is cited by
-
A second-generation M1-polarized CAR macrophage with antitumor efficacy
Nature Immunology (2024)
-
Therapeutic targeting of tumour myeloid cells
Nature Reviews Cancer (2023)
-
A lncRNA from an inflammatory bowel disease risk locus maintains intestinal host-commensal homeostasis
Cell Research (2023)
-
PD-L1 blockade in combination with carboplatin as immune induction in metastatic lobular breast cancer: the GELATO trial
Nature Cancer (2023)
-
METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer
Molecular Cancer (2023)