Abstract
Genotoxic DNA double-strand breaks (DSBs) can be repaired by error-free homologous recombination (HR) or mutagenic non-homologous end-joining1. HR supresses tumorigenesis1, but is restricted to the S and G2 phases of the cell cycle when a sister chromatid is present2. Breast cancer type 1 susceptibility protein (BRCA1) promotes HR by antagonizing the anti-resection factor TP53-binding protein 1(53BP1) (refs. 2,3,4,5), but it remains unknown how BRCA1 function is limited to the S and G2 phases. We show that BRCA1 recruitment requires recognition of histone H4 unmethylated at lysine 20 (H4K20me0), linking DSB repair pathway choice directly to sister chromatid availability. We identify the ankyrin repeat domain of BRCA1-associated RING domain protein 1 (BARD1)—the obligate BRCA1 binding partner3—as a reader of H4K20me0 present on new histones in post-replicative chromatin6. BARD1 ankyrin repeat domain mutations disabling H4K20me0 recognition abrogate accumulation of BRCA1 at DSBs, causing aberrant build-up of 53BP1, and allowing anti-resection activity to prevail in S and G2. Consequently, BARD1 recognition of H4K20me0 is required for HR and resistance to poly (ADP-ribose) polymerase inhibitors. Collectively, this reveals that BRCA1–BARD1 monitors the replicative state of the genome to oppose 53BP1 function, routing only DSBs within sister chromatids to HR.
Similar content being viewed by others
Main
Histone H4 lysine 20 (H4K20) methylation oscillates during the cell cycle, with major implications for chromosome replication, condensation and stability7. In the G1 phase, nucleosomes are fully methylated at H4K20, with >80% carrying H4K20 di-methylation (H4K20me2) (ref. 6). During S phase, new histone H4 unmethylated at K20 (H4K20me0) are incorporated on newly synthesized DNA and mixed in a 1:1 ratio with old nucleosomes methylated at H4K20. H4K20me0 thereby marks the post-replicative state of a genomic locus and thus the presence of a sister chromatid until G2/M6, when a surge of SET domain-containg protein 8 (SET8) methyltransferase activity catalyses mono-methylation of H4K20 (H4K20me1), which is subsequently converted to di- and tri-methylation by suppressor of variegation 4-20 homologue 1/2 (SUV4-20h1/2)6,7. The cell exploits this to regulate the recruitment of DNA repair factors. The TONSL (Tonsoku-like protein)–MMS22L (protein MMS22-like) homologous recombination (HR)8 complex reads H4K20me0 via its ankyrin repeat domain (ARD) to direct its function in DNA repair protein RAD51 homologue 1 (RAD51) loading9 to collapsed replication forks and DNA double-strand breaks (DSBs) in post-replicative chromatin6. Conversely, the non-homologous end-joining (NHEJ)-promoting factor TP53-binding protein 1 (53BP1) recognizes H4K20me1/2 (ref. 10) present on old histones throughout the cell cycle6. 53BP1 accumulation at DSBs is reduced in post-replicative chromatin11,12,13, correlating with the replication-dependent dilution of H4K20me1/2 (refs. 6,12,13). However, H4K20me1/2 dilution cannot explain 53BP1 suppression and the shift to HR, as breast cancer type 1 susceptibility protein (BRCA1) is required to antagonize 53BP1 accumulation at DSBs in S/G2 (refs. 11,12). DNA damage-induced ubiquitylation by RING finger proteins 8 (RNF8) and 168 (RNF168) is also required for both 53BP1 and BRCA1 recruitment14, but these signalling pathways are not cell-cycle specific14. The crucial question of how BRCA1 specifically recognizes post-replicative chromatin thus remains a missing part in the puzzle to understand DSB repair pathway choice.
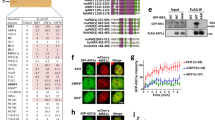
Because H4K20me0 directly marks sister chromatid availability, we set out to comprehensively explore its function. Using an unbiased quantitative proteomic strategy to identify proteins specifically recognizing nucleosomes carrying H4K20me0, we identified almost exclusively three post-replication DNA repair complexes; a BRCA1– BARD1 (BRCA1-associated RING domain protein 1)-containing complex involved in HR3; the RAD18 (an E3 ubiquitin-protein ligase)–SLF1 (SMC5–SMC6 complex localization factor 1)–SLF2 complex implicated in interstrand cross-link repair15; and TONSL–MMS22L, as expected6 (Fig. 1a). Consistent with previous work7,16, leucine-rich repeat and WD repeat-containing protein 1 (LRWD1), origin recognition complex subunit 1 (ORC1), ORC2 and ORC3 were specifically enriched on nucleosomes carrying H4K20me2 (Fig. 1a). Intriguingly, BARD1 and SLF1 both contain ARDs with high similarity to the TONSL ARD that recognizes H4K20me0 (ref. 6) (Fig. 1b and Supplementary Fig. 1), and the reported structure of the BARD1 ARD17 revealed that the histone H4-binding interface6 is structurally conserved (Fig. 1b). BARD1 and SLF1 showed a clear preference for H4K20me0 over H4K20me2 marked nucleosomes in pull-down experiments (Fig. 1c) and, importantly, mutation of three residues predicted to bind H4K20me0 (ARD 3A) based on TONSL homology abrogated nucleosome binding by SLF1 and BARD1 (Fig. 1b,d). These data identify two new H4K20me0 readers and argue for a general role of H4K20me0 in promoting post-replication DNA repair. In particular, recognition of H4K20me0 by BARD1 could provide a mechanism for BRCA1 recruitment to post-replicative chromatin, and directly link induction of HR to sister chromatid availability. Here, we explore this hypothesis.
a, Pull-downs from HeLa nuclear extracts with differentially modified biotinylated recombinant di-nucleosomes analysed by SILAC-based mass spectrometry. b, Left, alignment of the TONSL ARD (amino acids 528–626), BARD1 ARD (amino acids 427–546) and SLF1 ARD (amino acids 806–903). Right, overlay of the BARD1 ARD17 structure with the TONSL ARD from the TONSL–MCM2–H3–H4 structure6. Asterisks indicate the amino acids mutated in ARD 3A in d. c, Pull-downs with modified biotinylated di-nucleosomes in HeLa nuclear extracts (representative of three biological replicates). d, Pull-downs with biotinylated di-nucleosomes in nuclear extracts from HeLa cells expressing either wild-type or ARD 3A mutant forms of BARD1 or SLF1 (representative of three (BARD1) and two (SLF1) biological replicates). Unprocessed blots for c and d are presented in Supplementary Fig. 5.
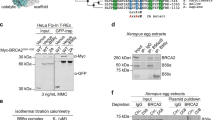
As H4K20me0 marks post-replicative chromatin irrespective of DNA lesions6, BRCA1–BARD1 might bind post-replicative chromatin in unperturbed cells. Indeed, we found progressive accumulation of the BRCA1–BARD1 complex on chromatin during the S phase mirroring H4K20me0 incorporation6 (Fig. 2a and Supplementary Fig. 2a,b), and this was dependent on H4K20me0 recognition by the BARD1 ARD (Fig. 2b and Supplementary Fig. 2c–e). BARD1 localized to 5-ethynyl-2’-deoxyuridine (EdU) pulse-labelled replication foci in the early S phase, but co-localization decreased in the mid- and late S phase (Fig. 2c). With continuous EdU labelling of post-replicative DNA, BARD1 and EdU co-localization remained high throughout the S phase (Fig. 2c), consistent with the recruitment of BARD1 via H4K20me0 present at sites of ongoing replication, as well as in post-replicative chromatin6. Depletion of SET8, which is responsible for methylation of H4K20 in G2 and mitosis, increased H4K20me0 levels and augmented BRCA1–BARD1 binding to chromatin in G2 and G1 in a manner dependent on H4K20me0 recognition by the BARD1 ARD (Fig. 2d and Supplementary Fig. 2f–h). SET8-depleted cells showed reduced NHEJ and elevated rates of HR and single-stranded DNA annealing18, but due to pleiotropic defects such as chromatin decompaction and DNA damage, this system was unsuitable for testing the role of H4K20me0 recognition in supporting BRCA1–BARD1-dependent HR. Taken together, our results establish ARD-dependent recognition of H4K20me0 as a mechanism for general pre-lesion recruitment of BRCA1–BARD1 to post-replicative chromatin, irrespective of DNA damage.
a, Top, high-content microscopy of chromatin-bound BARD1 and BRCA1 in pre-extracted U-2 OS cells pulsed with EdU. Bottom, cell-cycle stages were defined by gating on EdU and DAPI (top right). Mean ± s.d. intensities are shown relative to G1 levels (n = 3 independent experiments; from left, P = 0.0203, 0.0057, 0.0076 and 0.0170 for BARD1 (left), and 0.0055, 0.0315, 0.0251 and 0.0821 (NS, not significant) for BRCA1 (middle), ratio paired two-sided t-test). Data points represent the means of >173 cells. The correlation between BARD1 and BRCA1 mean intensities (lower right) is shown with Pearson’s R2 correlation (P < 0.0001). Representative cells are shown. Scale bars: 10 μm. b, Top, High-content microscopy of chromatin-bound Flag-HA-BARD1 in U-2 OS cells inducible for Flag-HA-BARD1 wild type or ARD 3A. Bottom, mean ± s.d. intensities are shown relative to wild type in G1 levels (n = 3 independent experiments; from left, P = 0.0214, 0.0036, 0.0220, 0.0204 and 0.0074, ratio paired two-sided t-test). Data points represent the means of >254 cells. c, Co-localization of chromatin-bound BARD1 and EdU in cells pulsed with EdU (left) or released into the S phase in the presence of EdU (right). S-phase stages were defined by EdU, DAPI and MCM2 staining6. Schematics illustrate the expected results based on H4K20me0 distribution6. Red lines and error bars represent means ± s.d. From left, n = 28, 22 and 18 (left), and 18, 25 and 22 cells (right). Representative cells are shown. Data were derived from two independent experiments. ****P < 0.0001, Mann–Whitney U-test (two-sided). NS results from left: P = 0.1392 and 0.4494. Scale bars: 15 μm. d, BARD1 and BRCA1 chromatin binding in the G1 (left) and G2 phase (right) in SET8-depleted cells. Cells inducible for siRNA-resistant Flag-HA-BARD1 wild type or ARD 3A were treated with BARD1 and SET8 siRNAs and analysed as in a. Mean ± s.d. intensities in SET8 and BARD1 co-depletion cells over BARD1 depletion alone are shown (n = 3 independent experiments; from left, P = 0.0048 and 0.0021 (for G1), and 0.013 and 0.0009 (for G2), ratio paired two-sided t-test). Data points represent the means of >206 cells.
Next, we analysed the role of H4K20me0 binding by BARD1 ARD in DNA DSB repair. Wild-type BARD1 recruitment to ionizing irradiation-induced DSBs increased during the S phase and reached a maximum in the late S phase before dropping in G2/M (Fig. 3a), mirroring H4K20me0 occupancy6 (Supplementary Fig. 2a) and recruitment of other HR factors11,19. Importantly, mutation of the BARD1 ARD ablated recruitment of both BARD1 and BRCA1 to DSBs in the S phase (Fig. 3a,b). Of note, the BARD1 wild type and ARD mutants were expressed at similar levels across the cell cycle and were proficient for BRCA1 stabilization via the RING domain dimerization3 (Supplementary Fig. 2c,e). Furthermore, we found that ARD recognition of H4K20me0 was as important as RNF8-dependent ubiquitination14 for BRCA1 accumulation at DSBs (Supplementary Fig. 3a), indicating that BRCA1–BARD1 recruitment requires multivalent interactions with chromatin at DSBs, similar to 53BP1 (ref. 20). Our finding that H4K20me0 is essential for BRCA1–BARD1 recruitment explains why forced methylation of H4K20 by non-degradable SET8 blocks BRCA1 recruitment and RAD51 loading at DSBs in the S phase12. Collectively, this establishes H4K20me0 recognition as the critical mechanism underlying specific recruitment of BRCA1 to DSBs in the S phase, and assigns a primary function to the BARD1 ARD.
a, Top, high-content microscopy of Flag-HA-BARD1 accumulation at ionizing irradiation-induced DNA DSBs. U-2 OS cells expressing Flag-HA-BARD1 wild type or ARD 3A were pulsed with EdU, irradiated with 1 Gy and, 45 min later, pre-extracted and analysed by immunofluorescence. Bottom, mean ± s.d. foci numbers are shown relative to the BARD1 wild type in mid-S phase. S-phase stages are defined as in Fig. 2a (n = 3 independent experiments). Data points represent the means of >381 cells. Scale bars: 10 μm. b,c, BRCA1 (b) and 53BP1 (c) accumulation at ionizing irradiation-induced DSBs in U-2 OS cells treated with control or BARD1 siRNAs and induced with TET to express siRNA-resistant Flag-HA-BARD1 wild type or ARD 3A. Cells were analysed as in a, and late S phase cells are shown. Means are shown as small ‘+’ signs, and whiskers indicate the 10th–90th percentiles. From left in b, n = 1,068, 723, 1,224, 896, 1,420, 627, 902 and 597 cells. From left in c, n = 1,068, 723, 1,224, 896, 1,420, 627, 902 and 597 cells. d, BRCA1 and 53BP1 foci in single cells in the late S phase were analysed as in b and c. For the BARD1 wild type, n = 896. For BARD1 ARD 3A, n = 597. e, High-content microscopy of single-stranded DNA measured by BrdU staining under non-denaturing conditions in U-2 OS cells treated with siRNA and TET as in c. Cells with high (top 15%, left) and low (bottom 15%, right) H4K20me0 levels (defined by gating on mean intensity) are shown. Cells were fixed 3 h after ionizing irradiation (3 Gy). Means are shown as small ‘+’ signs, and whiskers indicate the 10th–90th percentiles. From left, n = 503, 442, 371, 312, 474, 269, 381 and 330 (left), and 617, 641, 316, 237, 540, 495, 277 and 194 cells (right). f, High-content microscopy of ionizing irradiation-induced RAD51 foci analysed as in b. Means are shown as small ‘+’ signs, and whiskers indicate 10th–90th percentiles. From left, n = 310, 384, 327, 326, 329, 295, 321 and 331 cells.
A key essential activity of BRCA1 is to oppose the anti-resection activity of 53BP1 (refs. 4,5,11,12) to direct DSBs towards HR. 53BP1 accumulation at DSBs is dependent on recognition of H4K20me1/2 by its Tudor domain and RNF168-dependent mono-ubiquitination of histone H2A lysine 15 (ref. 14). However, BRCA1 can prevent 53BP1 access to repair sites11,12. We therefore hypothesized that BARD1 recognition of H4K20me0 might target BRCA1–BARD1-dependent exclusion of 53BP1 to post-replicative chromatin. BRCA1 foci dominated in the mid and late S phase in the presence of the BARD1 wild type as expected11, but, remarkably, this pattern was reversed by the BARD1 ARD mutant (Supplementary Fig. 3b). The BARD1 ARD mutant also completely failed to suppress 53BP1 accumulation at DSBs in BARD1-depleted S-phase cells (Fig. 3c,d). This switch from BRCA1–BARD1 to 53BP1 dominance ablated DNA end resection (Fig. 3e) and RAD51 loading (Fig. 3f), indicating that BARD1 ARD mutant cells cannot route DSBs to HR. These results show that BARD1 ARD recognition of H4K20me0 is required to recruit BRCA1 and curb 53BP1 anti-resection activity at DSBs in the S phase.
To address the implications for HR directly, we took advantage of a HR reporter carrying two differentially mutated green fluorescent protein (GFP) genes oriented as direct repeats (DR-GFP)21 using both U-2 OS cells depleted for endogenous BARD1 and a BARD1-null mouse mammary carcinoma cell line22. Mutation of the ARD ablated BARD1 function in HR in both systems (Fig. 4a,b). Consistent with a role of the H4K20me0 recognition in antagonizing 53BP1, depletion of 53BP1 partly restored HR in cells expressing the ARD mutant (Supplementary Fig. 3c). Moreover, mutation of the H4K20me0 recognition site was as detrimental to HR as deletion of the full ARD or BRCA1 carboxy (C) terminus (BRCT) domain previously shown to be essential for HR22 (Fig. 4b and Supplementary Fig. 3d), underscoring that the essential function of the ARD in HR is H4K20me0 recognition. In these experiments, we included mouse BARD1 as a control and found that it rescued HR activity (Supplementary Fig. 3d), although N470, predicted to bind H4K20me0 (Fig. 1b), is not conserved in mice. This suggested that the H4K20me0 reader domains in BARD1 and TONSL differ with respect to some H4 tail interactions. Taking advantage of the high-resolution structures of the TONSL ARD–H3–H4 (ref. 6) complex and the BARD ARD17, we used the Rosetta modelling package to exhaustively model the structure of the BARD1 ARD–H3–H4 complex. The best-scoring structures predicted a repositioning of H4 R17, reducing the contacts between the H4 tail and BARD1 N470 as compared to N571 in TONSL (Supplementary Fig. 4a,b). All other interactions were highly similar between the two reader domains—in particular, the contacts between BARD1 E467 and D500 with H4 H18 and the contacts between BARD1 E429, D458 and E467 with H4K20 were conserved (Supplementary Fig. 4a,b). Consistent with this binding model, residues E467 and D500, but not N470, were critical for BARD1 chromatin binding and recruitment to ionizing irradiation-induced DSBs (Supplementary Fig. 4c,d). These key acidic residues diverge from the consensus ankyrin repeat17, but are conserved across species in BARD1, TONSL and SLF1 (Supplementary Fig. 4e), explaining the specialization of these ARDs for H4K20me0 recognition.
a, Mean ± s.d. HR efficiency measured in DR-GFP U-2 OS cells treated with control or BARD1 siRNA and complemented with siRNA-resistant BARD1 wild type, ARD 3A or empty vector. The HR efficiency is shown as the percentage of control siRNA (n = 3 independent experiments). From left, P = 0.29 and 0.0016, ratio paired two-sided t-test. b, Mean ± s.d. HR efficiency measured in BARD1-null mouse mammary carcinoma cells carrying a DR-GFP reporter and complemented as indicated. The HR efficiency relative to empty vector is indicated above each bar (n = 3 independent experiments). c, BARD1 ARD mutations identified by cancer genome sequencing (COSMIC, https://cancer.sanger.ac.uk/cosmic; TCGA Research Network, https://cancergenome.nih.gov/). Mutations H459Y, D458V and E467K (red) are predicted to disturb H4 binding, as the residues are directly involved in H4 binding. Mutations D495Y, K503T, T534V and K540T (orange) are prone to affect the structural integrity of BARD1 ARD since the wild-type residues are involved in intramolecular hydrogen bonding. Mutations buried in the core (A435V, L447F, L480S, L498I and P530S; dark green) might affect the structure of the ARD and thus indirectly disrupt H4 binding. Surface-exposed mutations (light green) are less likely to affect ARD structure and H4 binding. d, Mean ± s.d. sensitivity to olaparib analysed by colony formation in siRNA-treated U-2 OS cells induced for the expression of siRNA-resistant Flag-HA-BARD1 wild type or ARD 3A (n = 3 independent experiments). e, HR efficiency measured in U-2 OS cells carrying a DR-GFP reporter, transfected with the indicated BARD1 constructs or empty vector along with an I-SceI expression vector (n = 4 independent experiments; *P = 0.0136, ratio paired two-sided t-test). Bars indicate mean ± s.d. f, Colony formation after olaparib (1 μM) treatment of U-2 OS cells induced to express Flag-HA-BARD1 wild type and ARD 3A by TET (mean, n = 3 independent experiments; from left, P = 0.9865 and 0.0013, ratio paired two-sided t-test).
Many mutations in the BARD1 ARD have been identified in cancer, and our structural model pinpoints those with the potential to disrupt H4K20me0 binding and promote carcinogenesis by crippling HR (Fig. 4c). HR deficiency is exploited in the treatment of tumours with BRCA1 and BRCA2 mutations23 as it sensitizes tumour cells to the poly (ADP-ribose) polymerase inhibitors (PARPi). Defects in H4K20me0 recognition are therefore expected to sensitize cells to PARPi treatment. Consistent with this prediction, a functional H4K20me0 recognition interface was required for BARD1 to rescue sensitivity to PARPi in BARD1-depleted cells (Fig. 4d). The BARD1 ARD mutant even impaired HR and sensitized cells to PARPi in a dominant negative manner (Fig. 4e,f), underscoring that H4K20me0 recognition is essential to BRCA1–BARD1 function in HR.
A role for the BARD1–BRCA1 E3 ubiquitin ligase activity in promoting HR was also recently reported and proposed to stimulate nucleosome remodelling events that inhibit 53BP1-dependent chromatin binding at DSB sites24. We therefore sought to determine the relative importance of H4K20me0 recognition in supporting BRCA1–BARD1-dependent HR over BARD1-dependent enzymatic E3 ubiquitin ligase activity. We generated a human conditional BARD1 knockout cell line by tagging both alleles of BARD1 with a C-terminal auxin-induced degron (AID)25, BARD1AID/AID (Fig. 5a), and then stably expressed BARD1 wild type or mutant transgenes. In BARD1AID/AID HCT116 cells, doxycycline-induced expression of a OsTIR1 transgene, integrated in the adeno-associated virus integration site 1 (AAVS1) locus, allowed for efficient and stable depletion of endogenous BARD1 following the addition of auxin indole-3-acetic acid (IAA; Fig. 5a). IAA-induced degradation of BARD1 resulted in BRCA1 destabilization (Fig. 5a) and PARPi hypersensitivity (Fig. 5b). The destabilization of BRCA1 was equally bypassed in cells expressing wild-type and mutant BARD1 transgenes (Fig. 5c). In addition to the recently reported BARD1 R99E RING domain mutant and BARD1 ARD 3A, we included a BARD1 BRCT-domain mutant (K619A)26 that abrogates poly (ADP-ribose)-dependent recruitment of BARD1 to stalled replication forks but is proficient for HR in mice27. In control (glutathione S-transferase-expressing) BARD1AID/AID cells, IAA treatment resulted in an acute hypersensitivity to PARPi (Fig. 5d). This hypersensitivity was completely reversed by the expression of wild-type BARD1, or the BARD1R99E and BARD1K619A mutants (Fig. 5d). In contrast, BARD1 ARD 3A expression failed to suppress PARPi sensitivity (Fig. 5d). These data indicate that ARD-mediated H4K20me0 binding represents a prime function for BARD1 in HR, while the E3-ligase activity and poly (ADP-ribose) binding are dispensable, consistent with these activities not being required for tumour suppression in mice27,28.
a, Western blot analysis of BARD1AID/AID HCT116 cells at indicated time points following the addition of doxycycline to stimulate OsTIR1 expression, and auxin (IAA) to induce OsTIR1-dependent BARD1-AID proteolysis. Representatives of two biological replicates are shown (see Supplementary Fig. 5 for unprocessed blots). Asterisks indicate non-specific bands. b, IAA addition hypersensitizes BARD1AID/AID HCT116 cells to olaparib. Cells were seeded in the presence of doxycycline. IAA and olaparib were added 1 d later and left for 7 d before staining with crystal violet. Representatives of two biological replicates are shown. c,d, Analysis of BARD1 ARD function in PARPi resistance, as defined in BARD1AID/AID HCT116 cells. After 24 h in doxycycline, the indicated cell lines were treated with IAA or carrier (dimethyl sulfoxide (DMSO)) before immunoblotting whole-cell lysates (c) or measuring growth in the presence of olaparib (d), as indicated. In c, western blots show equal stabilization of endogenous BRCA1 by the expression of wild-type and all mutant BARD1 transgenes, but not the control (glutathione S-transferase (GST)) transgene. Representatives of two biological replicates are shown (see Supplementary Fig. 5. for unprocessed blots). */eBARD1 indicates a weak unspecific band and the size of exogenous BARD1. In d, mean ± s.d. olaparib sensitivity was determined by resazurin cell viability assay 7 d following incubation with olaparib (a twofold dilution range from 7.8 nM–2 μM) in the presence (left) or absence of IAA (right) (n = 3 independent experiments). e, Schematic illustrating how BARD1 recognition of H4K20me0 licenses DSBs in sister chromatids for HR. BRCA1–BARD1 identifies post-replicative chromatin by recognition of H4K20me0 on new histones, thereby enabling the complex to oppose 53BP1 function when a sister chromatid can be used for homology-directed error-free repair of DSBs.
How cells switch from mutagenic NHEJ to error-free HR during S and G2 has been a long-standing question. Our results provide a simple solution in which ARD recognition of H4K20me0 triggers BRCA1–BARD1 recruitment specifically to post-replicative chromatin to antagonize 53BP1 and promote resection only when a sister chromatid is available for HR (Fig. 5e). This pre-lesion cis-acting mechanism acts at the most upstream point in repair pathway choice before cell-cycle regulation of end resection29 and recruitment of BRCA230, as well as H4K20me0-mediated RAD51 loading via TONSL–MMS22L6. We propose that two complementary mechanisms control HR in mammalian cells—sister chromatid sensing via H4K20me0 (this work and ref. 6) and cell-cycle regulation2, which is well established at many levels. Consistent with this, HR is strongly impaired when the deposition of new histones and thus marking of post-replicative chromatin by H4K20me0 is attenuated31. Recognition of H4K20me0 on new histones by BARD1, contrary to recognition of H4K20me1/2 on old histones by 53BP1, explains the specific recruitment of BRCA1 to DSBs in S/G2, including the competition between pro- and anti-resection activities at lesions in S/G2 chromatin32 where H4K20me0 and H4K20me1/2 marked nucleosomes coexist. At DNA break sites, H4K20 binding is integrated with additional signals, as both 53BP1 (ref. 20) and BRCA1–BARD1 (refs. 33,34,35 and this work) rely on multivalent binding for accumulation (including RNF8- and RNF168-dependent ubiquitination14). This cross-talk between the pathways with respect to ubiquitin signalling remains enigmatic but suggests a constant competition for chromatin binding between 53BP1 and BRCA1–BARD1 that converges at DSBs and is tuned to the replicative state of a genomic locus by H4K20 methylation status. Competition for ubiquitin binding at DSBs may also explain the futile recruitment of BRCA1 to DSBs in G1 cells that lack 53BP1 (ref. 36). In the presence of 53BP1, recognition of H4K20me0 provides BRCA1–BARD1 with an advantage in post-replicative chromatin, tipping the balance towards 53BP1 exclusion and induction of HR, which is then executed by downstream cell-cycle-regulated factors. Our data argue that ARD recognition of H4K20me0 mediates an essential HR function of BARD1, providing molecular insight into the function of the highly conserved ARD, and identifying the enigmatic HR function of BARD1 beyond BRCA1 stabilization37. Our work explains how the availability of a sister chromatid substrate for HR directs repair pathway choice, and the druggability of histone reader domains38 suggests that this mechanism can be exploited for targeting BRCA1 in cancers addicted to HR.
Methods
Preparation of recombinant histones
Recombinant human core histone proteins were expressed in Escherichia coli BL21(DE3)/RIL cells from pET21b(+) (Novagen) vectors, and purified by denaturing gel filtration and ion exchange chromatography, essentially as described39.
Preparation of truncated histone H4 for native chemical ligations
Truncated human H4Δ1–28I29C protein for ligations of modified histone H4 was expressed from a pET24b(+) vector (Novagen) in E. coli BL21(DE3)/RIL cells. The insoluble protein was extracted from inclusion bodies with unfolding buffer (20 mM Tris (pH 7.5), 7 M guanidine hydrochloride and 100 mM dithiothreitol (DTT)) for 1 h at room temperature, and the cleared supernatant was loaded on a Sephacryl S-200 gel filtration column (GE Healthcare) in SAU-1000 (20 mM sodium acetate (pH 5.2), 7 M urea, 1 M NaCl and 1 mM ethylenediaminetetraacetic acid (EDTA)) without any reducing agents. H4Δ1–28I29C-containing fractions were combined and further purified on a reversed-phase chromatography column (PerkinElmer Aquapore RP-300 (C8) 250 mm × 4.6 mm i.d.) using a gradient of 0–65% B (buffer A: 0.1% trifluoroacetic in water; B: 90% acetonitrile and 0.1% trifluoroacetic) over 20 column volumes. Fractions containing pure H4Δ1–28I29C were pooled and lyophilized.
Native chemical ligation
Native chemical ligations were carried out in 550 μl of degassed ligation buffer (200 mM KPO4, 2 mM EDTA and 6 M guanidine hydrochloride) containing 1 mg of modified/unmodified H4 peptides spanning amino acids 1–28 and containing a C-terminal thioester (Cambridge Peptides), 4 mg of truncated H4Δ1–28I29C, 20 mg 4-mercaptophenylacetic acid and 25 mg Tris(2-carboxyethyl)phosphine (TCEP) as a reducing agent at a pH of 7.5. The reactions were incubated overnight at 40 °C and quenched by the addition of 60 μl 1 M DTT and 700 μl 0.5% acetic acid. After precipitation clearance by centrifugation, the ligation reactions were directly loaded and purified on a reversed-phase chromatography column (PerkinElmer Aquapore RP-300 (C8) 250 mm × 4.6 mm i.d.) using a gradient of 35–45% B (buffer A: 0.1% trifluoroacetic in water; B: 90% acetonitrile and 0.1% trifluoroacetic) over 10 column volumes. Positive fractions containing ligated full-length histone H4 were combined and directly lyophilized.
Nucleosome assembly
Histone octamers were refolded from the purified histones and assembled into nucleosomes with biotinylated DNA via salt deposition dialysis as previously described39. Biotinylated nucleosomal DNAs containing two 601 nucleosome positioning sequences separated by a 50 base pair (bp) linker (di-nucleosomes) were prepared as described39. Di-nucleosomes were assembled in the presence of mouse mammary tumour virus A (MMTV A) competitor DNA and a slight excess of octamers to ensure saturation of the 601 repeats. The reconstituted di-nucleosomes were then immobilized on Streptavidin Sepharose High Performance beads (GE Healthcare) via the biotinylated DNA, washed to remove MMTV A competitor DNA and MMTV A nucleosomes, and directly used for stable isotope labeling with amino acids in cell culture (SILAC) nucleosome affinity purification (SNAP). Correct assembly and immobilization of nucleosomes was verified by native polyacrylamide gel electrophoresis.
SNAP
SILAC-labelled nuclear extracts were prepared from HeLa S3 cells as previously described39. For each pull-down, nucleosomes corresponding to 12.5 μg of octamer were immobilized on 10 μl Streptavidin Sepharose High Performance beads (GE Healthcare) in the final reconstitution buffer (10 mM Tris (pH 7.5), 250 mM KCl, 1 mM EDTA and 1 mM DTT; supplemented with 0.1% NP-40) and then rotated with 0.5 mg HeLa S3 SILAC-labelled nuclear extract in 1 ml of SNAP buffer (20 mM HEPES (pH 7.9), 150 mM NaCl, 0.2 mM EDTA and 10% glycerol) supplemented with 0.1% NP-40, 1 mM DTT and Protease Inhibitors cocktail (Roche) for 4 h at 4 °C. After 2 washes with 1 ml SNAP buffer + 0.1% NP-40, followed by two washes with 1 ml SNAP buffer without NP-40, the beads from both SILAC pull-downs were pooled. The supernatant was completely removed, and bound proteins were eluted by on-bead digestion.
On-bead digestion and peptide purification
The beads were resuspended in 50 μl of elution buffer (2 M urea, 100 mM Tris (pH 7.5) and 10 mM DTT) and incubated on a shaker (1,000 r.p.m.) at 25 °C for 20 min. Iodoacetamide (Sigma–Aldrich I1149) was added to a final concentration of 50 mM, and the sample was incubated on a shaker (1,000 r.p.m.) at 25 °C in the dark for 10 min. After digestion with 0.3 μg Trypsin (Promega V5113) for 2 h on a shaker (1,000 r.p.m.) at 25 °C, the supernatant was transferred to a new tube and was further digested with 0.1 μg Trypsin overnight at 25 °C. The digestion was stopped by adding 5.5 μl of 10% trifluoroacetic acid. Eluted peptides were purified on C18 stage-tips (Glygen 10–200 μl TopTips) following the manufacturer’s instructions and dried with a SpeedVac.
Mass spectrometry
Samples were loaded at 8 μl min−1 onto a trap column (Thermo Fisher Scientific Acclaim PepMap 100; 100 μm internal diameter, 2 cm length, C18 reversed-phase material, 5 μm diameter beads and 100 Å pore size) in 2% acetonitrile and 0.1% trifluoroacetic acid.
Peptides were eluted on line to an analytical column (Thermo Fisher Scientific Acclaim PepMap RSLC; 75 μm internal diameter, 25 cm length, C18 reversed-phase material, 2 μm diameter beads and 100 Å pore size) and separated using a flow rate of 250 nl min−1 and gradient conditions of: initial 5 min with 4% buffer B, then 90 min gradient 4–25% B, then 30 min gradient 25–45% B, then 1 min gradient 45–90% B, and finally 15 min isocratic at 100% B before returning to starting conditions for a 15 min equilibration (buffer A: 2% acetonitrile and 0.1% formic acid in water; B: 80% acetonitrile and 0.1% formic acid).
The Q Exactive instrument acquired full-scan survey spectra (m/z 300–1,650) at 70,000 resolution. An automatic gain control target value of 3 × 106 and a maximum injection time of 20 ms were used. The top 10 most abundant multiply charged ions were selected in a data-dependent manner, fragmented by higher-energy collision-induced dissociation, and data were collected over the range 200–2,000 m/z at 17,500 resolution. An automatic gain control target value of 1 × 105 with a maximum injection time of 120 ms was used. A dynamic exclusion time of 30 s was enabled.
Mass spectrometry data processing
Protein abundances from the Q Exactive mass spectrometer were quantified by MaxQuant 1.6.0.16 using 2-plex labeling (Arg10 and Lys8). The search was run against human UP000005640 proteome (version GCA_000001405.26) from UniProt. Search parameters were set up to allow variable oxidation on methionines and acetylation on amino termini, as well as fixed carbamidomethylation on cysteines. Tryptic peptides with up to two missed cleavages were considered for analysis, and tolerance settings were set to Orbitrap instrument. Co-fragmented peptide identification and matching between runs was enabled. For protein quantification, the minimum ratio count was set to 1, and remaining settings were left as default. The normalized heavy/light (H/L) ratios in proteinGroups.txt file output by MaxQuant were transformed to a log2 scale for further processing. Each protein in the dataset containing the ankyrin repeat-containing domain was identified using information from the InterPro superfamily IPR036770. Twenty-six proteins that were marked as either ‘potential contaminant’, ‘reverse’ or ‘only identified by site’ by MaxQuant were dropped from the dataset (that is, 1.81% of the data). A further 76 proteins (5.39% of the remaining data) were dropped because they had only a forward or reverse ratio, but not complete pairs. The data presented in Fig. 1a are zoomed in to log2[fold change] ratios below the value of 5, thus excluding 4 outlier proteins (SPTA1, STAG2, histone H3 and histone H4) that are not enriched in either nucleosome pull-down. The full proteomics list for H4K20me0/me2 nucleosome pull-downs is presented in Supplementary Table 1.
Di-nucleosome pull-down assay
Biotinylated di-nucleosomes carrying H4K20me0 or H4K20me2, or biotin only as a control, were mixed with MyOne T1 Streptavidin beads (Thermo Fisher Scientific) in Coupling Buffer (10 mM Tris-HCl, 1 mM EDTA and 250 mM KCl). Di-nucleosomes and beads were incubated for 1 h at 4 °C on a rotating shaker to allow the coupling and, subsequently, washed twice with SNAP buffer (20 mM HEPES, 150 mM NaCl, 0.2 mM EDTA, 10% glycerol and 0.1% NP-40). At the same time, 320–450 μg of HeLa cell nuclear extracts were incubated with 10 μg ml−1 Herring Sperm DNA (Sigma–Aldrich) in SNAP buffer (final volume: 900 μl) and allowed to rotate for 1 h at 4 °C. Inputs of 2% of the extracts were taken before diluting each sample with SNAP buffer. The extracts were then added to the washed di-nucleosome-coupled beads and incubated for 4 h, rotating at 4 °C. The beads were then washed with 4 × 900 μl Wash Buffer (20 mM HEPES, 500 mM NaCl, 0.2 mM EDTA, 10% glycerol and 0.25% NP-40, supplemented with 10 μg ml−1 Herring Sperm DNA) for 2 min at 4 °C. After the last wash, the beads were carefully dried and boiled in 20 μl 1× Laemmli sample buffer (LSB; 50 mM Tris–HCl pH 6.8, 100 mM DTT, 2% SDS, 8% glycerol, bromophenol blue). Pull-downs were visualized by western blotting after protein separation on a NuPAGE 4–12% Bis-Tris gradient gel (Thermo Fisher Scientific).
Cell culture, transfection and drug treatment
U-2 OS (a gift from J. Bartek), HeLa S3 (a gift from P. Nakatani) and HCT116 cells (a gift from I. Tomlinson) were grown in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% foetal bovine serum (HyClone), 1% penicillin/streptomycin and drugs for selection. Cells inducible for Flag-HA-BARD1 wild-type and ARD mutants were generated in Flp-In T-REx U-2 OS cells (Invitrogen) by transfection of pcDNA5/FRT/TO-Flag-HA-BARD1 plasmids with Lipofectamine 2000, according to the manufacturer’s protocol, and selection with hygromycin (200 μg ml−1). All cell lines were authenticated by western blotting and/or immunofluorescence. All cell lines used in this study tested negative for Mycoplasma contamination. Expression of Flag-HA-BARD1 was induced by the addition of 1 μg ml−1 of tetracycline (TET) for 24–48 h. For transient expression of Flag-HA-BARD1, expression plasmids were introduced by transfection with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol and cells harvested 24–48 h post-transfection. To establish a parental cell line expressing the E3 ubiquitin ligase OsTIR1, HCT116 male colorectal carcinoma cells were simultaneously transfected with a version pX330 (Addgene plasmid 42230) containing a locus-specific guide RNA targeting AAVS1 (5′-GGGGCCACTAGGGACAGGAT-3′) and pMK243 Tet-OsTIR1 (Addgene plasmid 72835) using FuGENE HD (Promega). Individual subclones were isolated and assessed for doxycycline inducible expression by western blot using an OsTIR1-specific antibody (a gift from M. Kanemaki at the National Institute of Genetics). Integration of Tet-OsTIR1 at the AAVS1 locus was confirmed by PCR. siRNA transfection was performed with RNAiMax reagent (Invitrogen) according to the manufacturer’s protocol. siRNA sequences (Sigma–Aldrich) were: siBARD1, 5′-AAGCUGUUGCCCAAUAUGGCU-3′; siRNF8, 5′-UGCGGAGUAUGAAUAUGAA-3′; and si53BP1, 5′-GAAGGACGGAGUACUAAUA-3′. The SET8 siRNAs have been described previously6.
AID tagging of endogenous BARD1 in Tet-OsTIR1 HCT116 cells
One day before transfection, 3.2 × 105 Tet-OsTIR1 HCT116 cells were seeded in a single well of a 6-well plate. 800 ng of pX330 containing a guide RNA specific to the C terminus of BARD1 (5′-TATAATATTCAGCTGTCAAG-3′) was combined with 600 ng of circular donor plasmid and transfected using FuGENE HD (Promega) according to the manufacturer’s protocol. After 48 h selection in blasticidin (10 μg ml−1), surviving cells were seeded at low density in a 10 cm dish for subcloning. Genomic DNA was isolated from individual clones following expansion, and biallelic integration of the AID tag and selection cassette was assessed by PCR with primers flanking the integration site (BARD1_IntegrationScreen_R: 5′-GCATTCATTTTATGTTTCAGGTTCAGG-3′; and BARD1_IntegrationScreen_F: 5′-CTGTTTGATGGATGCTACTTCTATTTG-3′). Individual alleles of promising clones were TA cloned into pJET 1.2 and sequenced to ensure clonal purity. Complementation of confirmed BARD1AID/AID clones was carried out by stable transduction using Lentivirus.
Plasmid construction
A fragment of BARD1 encompassing the ARD (amino acids 406-558) was synthetized by GenScript to contain silent mutations to generate siRNA resistance (5′ – AAGCTGcTGCCtAAcATGGCc – 3′) in combination with BARD1 ARD mutations (N470A, N470S, E467A, D500A and N470AE467AD500A (ARD 3A)). siRNA-resistant pcDNA5–FRT/TO–Flag–HA–BARD1 constructs used to generate inducible Flp-In U-2 OS cell lines were constructed as follows. The BARD1 406–558 fragment was amplified with left and right homology arms complementary to the BARD1 sequence outside the 406–558 fragment (Fw: 5′ – TAGTTCAAGTTACAGGCAAGTGATGTCTAG – 3′, Rev: 5′ – GCCCAGTGTTCATTACTGAGCAGTGGCTAG – 3′); a pCIN4–Flag–BARD1 plasmid (a kind gift from Richard Baer at the Institute of Cancer Genetics, Columbia University, New York) was amplified by inverse PCR amplification using primers that annealed immediately outside to the BARD1 406–558 fragment (Fw: 5′ – TAATGAACACTGGGCAGCGT – 3′, Rev: 5′ – CTGTAACTTGAACTACTTAATGTA – 3′). The two fragments were then combined by InFusion cloning (Clontech) before the full BARD1 cDNA was transferred to a pcDNA5–FRT/TO–Flag–HA vector and verified by sequencing. A donor plasmid for endogenous AID tagging of the BARD1 C terminus was constructed by overlapping fusion PCR of 3 fragments: (1) a left homology arm of 233 bp amplified from HCT116 genomic DNA using oligos BARD1_LHomArm_F (5′-GTGACTCAGACCATCAATACAG-3′) and BARD1_LHomArm_R (5′-TCTTTAGGACAAGCACTCTTCTCCTTGGCGCCTGCACCGCTGTCAAGAGGAAGCAACTCAAATGAC-3′); (2) a right homology arm of 257 bp amplified from HCT116 genomic DNA using oligos BARD1_RHomArm_R (5′-GCAATCCCAGCTTCTAAATGGTAAAC-3′) and BARD1_RHomArm_F (5′-ATTAGGTCCCTCGAAGAGGTTCACTAGGATCCGGTACCCAAGTTGCTTCCTCTTGACAGCTG −3′); and (3) an AID tag and blasticidin selection cassette amplified from pMK288 (Addgene plasmid 72826; PMID 27052166) using oligos BARD1_AIDCassette_F (5′-CGGTGCAGGCGCCAAGGAGAAGAGTG-3′) and BARD1_AIDCassette_R (5′-TGGGTACCGGATCCTAG-3′). All three fragments were fused in a PCR reaction containing BARD1_LHomArm_F and BARD1_RHomArm_R, and the fused product was ligated into pJET 1.2 by blunt end cloning following the CloneJET PCR Cloning Kit manufacturer’s protocol (Thermo Fisher Scientific).
Immunofluorescence, microscopy and ionizing irradiation
U-2 OS cells conditional for Flag-HA-BARD1 were grown on glass coverslips or 96-well plates and either directly fixed in 4% paraformaldehyde for 10 min or washed in cytoskeleton buffer (CSK; 10 mM PIPES pH 7, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2), pre-extracted after 5 min with CSK/0.5% Triton X-100 (on ice) and rinsed with CSK and phosphate buffered saline before fixation in 4% paraformaldehyde for 10 min. For high-content quantitative analysis, fluorescence images were acquired using an Olympus ScanR high-content microscope and processed on the ScanR analysis software. More than 5,000 cells per sample were analysed. Cell-cycle phases were gated on 4′,6-diamidino-2-phenylindole (DAPI) and EdU intensity. For co-localization analysis by deconvolution microscopy, images were acquired on a DeltaVision microscope with a 60× oil objective and analysed by SoftWoRx 6.5.2. Pearson’s coefficient correlation analysis was performed on single cells using SoftWoRx 6.5.2. Ionizing radiation treatment was given using an X-ray apparatus (Faxitron) calibrated to give 1 Gy min−1. For analysis of DNA end resection, 5-bromo-2’-deoxyuridine (BrdU) labelling and detection under non-denaturing conditions was performed as previously described12. Briefly, U-2 OS cells were pre-incubated with 30 μM BrdU for 24 h and then treated with ionizing irradiation (3 Gy). After 3 h recovery, cells were pre-extracted with nuclear extraction buffer (20 mM HEPES-NaOH (pH 7.5), 20 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% NP-40, 100 μM phenylmethanesulfonyl fluoride, 10 μM pepstatin, 10 μM leupeptin, 0.2 mM Na3VO4, 10 mM β-glycerophosphate and 5 mM NaF) on ice for 10 min and fixed with 4% formaldehyde/phosphate buffered saline on ice for 10 min.
HR assay
DR-GFP U-2 OS or BARD1-null mouse mammary carcinoma cells (18–09) were used as described previously21,22. Briefly, 2.5–5 × 105 cells well−1 were seeded on 6-well plates. After 24 h, the cells were transfected with 1 μg of I-SceI expression vector (pCBASce), 2 μg of BARD1 expression vector or empty vector and 1 μg of monomeric red fluorescent protein (mRFP) expression vector (pCS2–mRFP). 24 h after transfection, the cells were washed and re-plated on 100 mm dishes. Flow cytometric analysis to quantify the presence of GFP-positive cells was performed two days later on a FACSCalibur using CellQuest Pro software (BD Biosciences). For each sample, 30,000 cells were assessed, and the percentage of GFP-positive cells in RFP-positive cells was calculated as HR repaired cells. RFP-encoding vector was used to provide a control for transfection efficiency. For 53BP1 depletion, the siRNAs were transfected 24 h before plasmid transfection.
Clonogenic assay
U-2 OS inducible for the FLAG-HA-BARD1 wild type and ARD 3A mutant were seeded in technical triplicates of 300–10,000 cells in the presence or absence of TET. After 48 h, olaparib (Selleck Chemicals; S1060) was added for 72 h. Cells were then cultured in fresh medium for 7–15 d before fixation and staining with MeOH/Crystal Violet. For the complementation, BARD1 wild-type and ARD 3A mutant cells were transfected with siRNA and seeded with or without TET. The cells were trypsinized 24 h later and seeded in technical triplicates of 300–10,000 cells in the presence or absence of TET. After 24 h, olaparib was added for 24 h at the indicated concentrations. Cells were then cultured in fresh medium for 7–15 d before fixation and staining with MeOH/Crystal Violet. The colony formation efficiency was determined by manual colony counting, and normalized to the non-drug-treated control. Each data point represents a technical triplicate of seeded cells within each biological replicate.
Resazurin cell viability assay
Tet-OsTIR1 HCT116 cells (300 well−1) were seeded in triplicate in 96-well plates in the presence of doxycycline (1 mg ml−1) for each concentration of olaparib (Selleck Chemicals; S1060). Then, 24 h after plating, IAA was added to a final concentration of 1 mM. One hour after the addition of IAA, olaparib was added to indicated final concentrations. The medium was refreshed at 4 d. Seven days after the addition of olaparib, the medium was replaced with phenol red-free Dulbecco’s modified Eagle’s medium (10% foetal bovine serum and 1% penicillin/streptomycin) containing 10 mg ml−1 resazurin, and the plates were incubated for 2 h. The relative fluorescence intensity of each well was then measured using a BMG LABTECH CLARIOstar plate reader equipped with a 560 nm excitation and 590 nm emission filter set.
Antibodies
The following antibodies were used for western blot: BARD1 (Bethyl A300-263A; 1:500), BARD1 (Abcam ab64164; 1:500), SLF1 (Novus Biologicals NBP1-88358; 1:500), HA (HA.11, BioLegend 901513 (previously Covance MMS-101P); 1:500), H4K20me2 (Diagenode C15200205; 1:3000), GFP (Roche 11 814 460 001 (mixture of clones 7.1 and 13.1); 1:500), SET8 (Millipore, 06-1304; 1:1000), OsTIR1 (a gift from M. T. Kanemaki; 1:1000), Actin (Sigma–Aldrich A1978; 1:2000) and BRCA1 (Santa Cruz Biotechnology sc-6954, clone 9; 1:400). The following antibodies were used for immunofluorescence: BARD1 (Bethyl A300-263A; 1:500), BRCA1 (Santa Cruz Biotechnology sc-6954, clone 9; 1:400), 53BP1 (Santa Cruz Biotechnology H-300; 1:500), H4K20me1 (Abcam ab9051; 1:250 (validated in Supplementary Fig. 2g)), H4K20me2 (Diagenode C15200205; 1:250 (validated in Supplementary Fig. 2g)), H4K20me0 (Abcam ab227804; 1:10000 (validated in Supplementary Fig. 2g)), Phospho-H2A.X (S139), Cell Signaling Technology 2577; 1:1000), HA (Roche 1 867 423; 1:200), HA (BioLegend 901509; 1:100), BrdU (Eurobio ABC117-7513; 1:1000) and DNA replication licensing factor MCM2 (MCM2; BD Biosciences 610701; 1:150).
Modelling the structure of BARD1 bound to histone H4
The Rosetta macromolecular modelling package (www.rosettacommons.org), which has proven successful in a variety of protein design applications involving protein structure predictions (predictions of protein–protein and protein–peptide interfaces) was used to model the interaction of the histone H4 tail with BARD1. Due to the high structural similarity of TONSL (Protein Data Bank (PDB) ID: 5JA4) and BARD1 ARD (PDB ID: 3C5R) (main chain root-mean-square deviation = 0.96 Å), we chose to use the structure of TONSL in complex with the histone H3–H4 tetramer as a seed structure to model the complex of the histone H4 tail bound to BARD1. The starting structure for the Rosetta refinement of BARD1 bound to histone H4 was prepared from the structural superposition of the ARD domains of TONSL and BARD1. Analysis of the TONSL ARD and histone H4 interaction interface (PDB ID: 5JA4) suggested that the imidazole side chain of H18 (in H4) is positively charged, as it is located in a cavity between the negatively charged residues E568 and D604 that is conserved (E467 and D500) in the ARD domain of BARD1. A positively charged H18 was also supported by pKa calculations using the Rosetta protocol to calculate pKa values. As histidines in Rosetta are assumed to be neutrally charged by default, we prepared a parameter file to protonate both the epsilon and delta state of H18 to carry a net (+1) positive charge. The preliminary model of the BARD1 and H3–H4 complex was exhaustively refined using Rosetta 5.0.37 by subjecting it to 3,000 steps of full-atom structure relaxation (in the Rosetta force field), which samples side-chain and backbone conformational changes (similar to those suggested to underlie observed conformational heterogeneity in high-resolution crystal structures) to broadly sample the protein–protein interface and the conformational variability of the complex40. For each relax cycle, five rounds of repacking followed by gradient base minimization in torsion space (backbone φ/ψ and side chain χ torsional degrees of freedom) was performed until convergence (an absolute score change on minimization of less than one Rosetta energy unit). From all cycles performed, the best-scoring pose was selected to represent the structure of BARD1 bound to the histone H4 tail, as shown in Supplementary Fig. 4a.
Statistics and reproducibility
Statistics were analysed with GraphPad Prism 7. All statistics were evaluated by either independent or paired two-tailed Student’s t-test and two-tailed Mann–Whitney U-test. Pearson’s correlation and Student’s t-test were performed under the assumption of normality. The exact sample sizes (n) used to calculate statistics are provided in the figure captions. P values are provided in the figure legends and captions. Whiskers indicate 10th–90th percentiles, and outliers are plotted as individual points in box-and-whisker plots. All experiments were reproduced with similar results a minimum two times.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
The code used to analyse the mass spectrometry data is publicly available at GitHub (https://github.com/lukauskas/publications-nakamura-2018-snap-h4k20me2).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009281, and are presented in Supplementary Table 1. Unprocessed images of all gels and blots (Figs. 1c,d and 5a,c and Supplementary Fig. 2h) are provided in Supplementary Fig. 5. Source data for all graphs are provided in Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
References
Moynahan, M. E. & Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 (2010).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2016).
Prakash, R., Zhang, Y., Feng, W. & Jasin, M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7, a016600 (2015).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Zimmermann, M. & de Lange, T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 24, 108–117 (2014).
Saredi, G. et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature 534, 714–718 (2016).
Beck, D. B., Oda, H., Shen, S. S. & Reinberg, D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 26, 325–337 (2012).
Piwko, W., Buser, R. & Peter, M. Rescuing stalled replication forks: MMS22L-TONSL, a novel complex for DNA replication fork repair in human cells. Cell Cycle 10, 1703–1705 (2011).
Piwko, W. et al. The MMS22L-TONSL heterodimer directly promotes RAD51-dependent recombination upon replication stress. EMBO J. 35, 2584–2601 (2016).
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006).
Chapman, J. R., Sossick, A. J., Boulton, S. J. & Jackson, S. P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 125, 3529–3534 (2012).
Pellegrino, S., Michelena, J., Teloni, F., Imhof, R. & Altmeyer, M. Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin. Cell Rep. 19, 1819–1831 (2017).
Simonetta, M. et al. H4K20me2 distinguishes pre-replicative from post-replicative chromatin to appropriately direct DNA repair pathway choice by 53BP1-RIF1-MAD2L2. Cell Cycle 17, 124–136 (2018).
Uckelmann, M. & Sixma, T. K. Histone ubiquitination in the DNA damage response. DNA Repair (Amst.) 56, 92–101 (2017).
Raschle, M. et al. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348, 1253671 (2015).
Kuo, A. J. et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature 484, 115–119 (2012).
Fox, D. et al. Crystal structure of the BARD1 ankyrin repeat domain and its functional consequences. J. Biol. Chem. 283, 21179–21186 (2008).
Tuzon, C. T. et al. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep. 8, 430–438 (2014).
Karanam, K., Kafri, R., Loewer, A. & Lahav, G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 47, 320–329 (2012).
Wilson, M. D. et al. The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103 (2016).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999).
Laufer, M. et al. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J. Biol. Chem. 282, 34325–34333 (2007).
Jackson, S. P. & Helleday, T. Drugging DNA repair. Science 352, 1178–1179 (2016).
Densham, R. M. et al. Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23, 647–655 (2016).
Natsume, T., Kiyomitsu, T., Saga, Y. & Kanemaki, M. T. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016).
Li, M. & Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23, 693–704 (2013).
Billing, D. et al. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell 72, 127–139 (2018).
Shakya, R. et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334, 525–528 (2011).
Huertas, P., Cortes-Ledesma, F., Sartori, A. A., Aguilera, A. & Jackson, S. P. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 (2008).
Orthwein, A. et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015).
Huang, T. H. et al. The histone chaperones ASF1 and CAF-1 promote MMS22L-TONSL-mediated Rad51 loading onto ssDNA during homologous recombination in human cells. Mol. Cell 69, 879–892 (2018).
Ochs, F. et al. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 23, 714–721 (2016).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Escribano-Diaz, C. et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013).
Westermark, U. K. et al. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 23, 7926–7936 (2003).
Zaware, N. & Zhou, M. M. Chemical modulators for epigenome reader domains as emerging epigenetic therapies for cancer and inflammation. Curr. Opin. Chem. Biol. 39, 116–125 (2017).
Bartke, T. et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–484 (2010).
Conway, P., Tyka, M. D., DiMaio, F., Konerding, D. E. & Baker, D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 23, 47–55 (2014).
Acknowledgements
We thank J. Lukas for commenting on the manuscript, researchers at the Groth laboratory for fruitful discussions, R. Baer for sharing the BARD1 plasmid and BARD1-null mouse mammary carcinoma cells, M. Kanemaki for the OsTIR1 antibody, Y. Antoku for assistance with microscopy, and J. V. Johansen for help with statistical analysis. J.R.B. is funded by a Cancer Research UK Career Development Grant (C52690/A19270). Funding for T.B. was provided by the Medical Research Council (grant number MC_UP_1102/2) and European Research Council (ERC StG number 309952). S.L. was supported by a stipend from the Biotechnology and Biological Sciences Research Council. The Groth laboratory is supported by the Danish Cancer Society, Novo Nordisk Foundation, Lundbeck Foundation, European Research Council (ERC CoG number 724436), Independent Research Fund Denmark and Neye Foundation.
Author information
Authors and Affiliations
Contributions
K.N., G.S. and A.G. conceived the study. K.N. carried out the functional analysis. G.S. performed pull-downs with endogenous BARD1 and SLF1, and established tools and reagents. J.R.B. generated and carried out experiments with BARD1AID/AID cells. B.M.F. prepared materials for nucleosome pull-downs. N.V.N. prepared materials, carried out SILAC nucleosome pull-down experiments and analysed the data. T.E.B. and L.C.C. performed pull-downs with BARD1 and SLF1 mutants. P.A.F. carried out mass spectrometry measurements and analysed the mass spectrometry data. S.L. performed bioinformatics analyses of the mass spectrometry data. T.F. performed structural modelling. J.R.C., T.B. and A.G. supervised the project and analysed the data. G.S. and A.G. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
G.S. and A.G. are inventors on a filed patent application covering the therapeutic targeting of ARD interactions with H4K20me0 for cancer therapy. A.G. and T.F. are co-founders of Ankrin Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Domain architecture of TONSL, BARD1 and SLF1.
Domain boundaries are indicated. TPR, Tetratricopeptide Repeats. UBL, Ubiquitin-Like. LRR, Leucine Rich Repeats. RING, Really Interesting New Gene finger domain. BRCT, BRCA1 C-Terminus.
Supplementary Figure 2 ARD recognition of H4K20me0 recruits BRCA1–BARD1 to post-replicative chromatin.
a, High-content microscopy of H4K20me0/1/2 levels across the cell cycle. U-2-OS cells were pulsed with EdU, pre-extracted, fixed and processed for immunofluorescence. Cell cycle stages were defined by gating on EdU and DAPI as shown (right). Red bars indicate mean. b, One representative experiment from Fig. 2a is shown. Red bars indicate mean. From left, n = 3357, 2367, 3491, 800, 1721 and 3357, 2367, 3491, 800, 1721. c, Control for equal expression of Flag-HA-BARD1 WT and ARD 3A in inducible U-2-OS cells lines used in Fig. 2b, d, 3a–f. Cells induced or not for Flag-HA-BARD1 WT and ARD 3A expression with TET were fixed directly (without pre-extraction) and stained for HA. Mean HA-BARD1 intensity is indicated relative to HA-BARD1 WT intensity in TET treated G1 cells. Mean with S.D. is shown, n = 3. NS, not significant, ratio paired two-sided t-test. From left, P = 0.3004, 0.3867, 0.9878, 0.2141, 0.9006. d, One representative experiment from Fig. 2b is shown. Red bars indicate mean. From left, n = 3516, 2119, 2495, 953, 1265 and 4642, 2074, 2949, 1198, 1285. e, Control for total expression of BARD1 and BRCA1 in siRNA-treated U-2-OS cells inducible for Flag-HA-BARD1 WT and ARD 3A used in Fig. 2b, d, 3a–f. High-content microscopy of BARD1 and BRCA1 across the cell cycle in siRNA-treated U-2-OS cells induced or not with TET to express Flag-HA-BARD1 WT or ARD 3A. Mean with S.D. is shown, n = 3. Each data point represents the mean of >603 cells. IR treatment: 1 Gy, 2 h recovery before HA staining (right panel). NS, not significant, ratio paired two-sided t-test. f, One representative experiment from Fig. 2d is shown. Mean shown as ‘+’, whiskers indicate 10–90 percentile. From left, (G1) n = 1513, 466, 1203, 478, (G2) n = 685, 551, 626, 495. g, High content microscopy of H4K20me0/1/2 levels in G1 and G2 cells depleted for SET8 as in Fig. 2d. Red bars indicate mean. Cell cycle stages were defined by gating on EdU and DAPI as shown. h, Western blot analysis of H4K20me1/2 levels in SET8 depleted U-2-OS cells. Representative of 2 biological replicates. Unprocessed blots are presented in Supplementary Fig. 5.
Supplementary Figure 3 ARD recognition of H4K20me0 is required for recruitment of BRCA1–BARD1, exclusion of 53BP1 and HR at DSBs in S/G2.
a, Contribution of RNF8 and H4K20me0 recognition to BRCA1 recruitment at DSBs in late S. High-content microscopy of BRCA1 in siRNA-treated U-2-OS cells induced with TET to express Flag-HA-BARD1 WT or ARD 3A. Cells were fixed 1 h after treatment with IR (5 Gy). The number of BRCA1 foci is shown relative to the control siRNA in Flag-HA-BARD1 WT cells without TET induction. Late S phase cells are defined by gating on EdU and DAPI as in Fig. 2a. Mean with S.D. is shown, n = 3. b, The role of BARD1 ARD recognition of H4K20me0 in antagonizing 53BP1 accumulation. High-content microscopy of IR-induced BRCA1 and 53BP1 foci across the cell cycle in U-2-OS cells induced to express Flag-HA-BARD1 WT or ARD 3A. Left, single cell analysis of BRCA1 and 53BP1 foci and DAPI total intensity. Right, quantification of 4 independent experiments. Foci number is shown relative to BARD1 WT in mid S. Mean with S.D. is shown, n = 4. *P < 0.05, **P < 0.001. NS, not significant, ratio paired two-sided t-test. From left, P = 0.009, 0.0661, 0.0133, 0.0771, 0.0513, 0.0072, 0.0782, 0.0083, 0.0528, 0.0202. c, The HR defect in BARD1 ARD expressing cells is partly rescued by 53BP1 depletion. DR-GFP U-2-OS cells were siRNA treated as indicated and complemented with siRNA-resistant Flag-HA-BARD1 ARD 3A cDNA or empty vector before DR-GFP analysis. HR efficiency is shown relative to control siRNA. Mean with S.D. is shown, n = 3, from left, P = 0.0195 0.0106, ratio paired two-sided t-test. The gating strategy to identify transfected (RFP positive) and HR proficient (GFP positive) cells is shown on the right. d, Complementation of BARD1-null DR-GFP reporter cells with the indicated BARD1 cDNAs as in Fig. 4b, including mouse BARD1 and additional controls. The HR efficiency relative to empty vector (vector, pIRESpuro) is indicated above each bar. The BARD1 domain deletions carry a sequence divergence (Q406)23 from the currently annotated sequence (R406, UniProt ID: Q99728). Importantly, BARD1 Q406 (WT Q406) and BARD1 R406 (WT) were equally proficient in rescuing HR efficiency and for simplicity the sequence divergence is therefore not annotated in Fig. 4b, but indicated here: WT, BARD1 (R406); ARD 3A (R406); WT Q406 (Q406)23; ΔARD (Q406)23; ΔBRCT (Q406)23; mBARD1, mouse BARD1. Mean with S.D., n = 3.
Supplementary Figure 4 Structural modelling of the BARD1 H4K20me0 reader domain and analysis of single point mutations in the ARD.
a, b, Model of the BARD1 ARD structure (green) in complex with histone H4 (L22-K12) tail (yellow) refined by the Rosetta macromolecular modelling package using the 2.4 Å high-resolution crystal structure of TONSL ARD/H4K20me07 (PDBID 5ja4) as template (see method section for further details). The C-alpha Root-Mean-Square Deviation (RMSD) between the BARD1 ARD model in complex with H3-H4 and the BARD1 ARD crystal structure18 (PDBID 3c5r - in absence of H4) is 0.7 Å. In comparison, the main chain RMSD between the ARD domains of the crystal structure of BARD1 (PDBID 3c5r) and TONSL (PDBID rja4) is 0.9 Å. The hydrogen bonding network of predicted key interactions between the positively charged side chains of H18 and K20 (yellow sticks) and the negatively charged E429, D458, E467 and D500 (green sticks) are illustrated by yellow dashed lines. (b) Structural super positioning of BARD1 ARD/H4 (L22-K12) (green/yellow) with the crystal structure (PDBID 5ja4) of TONSL ARD/H4K20me0 (grey). Repositioning of R17 in H4 is indicated by the dashed black arrow. In the TONSL crystal structure R17 makes interactions with both the main and side chain carbonyl of N571. In the model of the BARD1 ARD in complex with H4, NH2 in the amide side chain of the nearby N504 (which is C608 in TONSL ARD) makes an intramolecular hydrogen bond to the backbone carbonyl of N470 resulting in a relocation of R17. The carbonyl oxygen of the N504 side chain is predicted to make a hydrogen bond interaction with the NH amide backbone atom of K16. c, d, Analysis of BARD1 ARD single amino acid mutants in U-2-OS cells induced to express Flag-HA-BARD1 WT or the indicated mutants. Mean with S.D., n = 3. (c) Total protein levels and chromatin-binding were analysed by high content microscopy in cells either directly fixed or pre-extracted before being processed for HA immunofluorescence. (d) High content microscopy of Flag-HA-BARD1 and BRCA1 IR-induced foci analysed 45 min after IR (3.5 Gy). All constructs carried the Q406 sequence divergence, as described in Supplementary Fig. 3d. e, Alignment of the ARDs of TONSL, BARD1 and SLF1 (from H. sapiens, M. musculus and X. tropicalis). The general ARD consensus sequence is indicated above. The arrows indicate the conserved acidic residues.
Supplementary information
Supplementary Information
Supplementary Figures 1–5 and legends for Supplementary Tables 1 and 2.
Supplementary Table 1
Full list of proteins identified by SILAC mass-spectrometry pull-down with H4K20me0 and H4K20me2 di-nucleosomes.
Supplementary Table 2
Statistics Source Data.
Rights and permissions
About this article
Cite this article
Nakamura, K., Saredi, G., Becker, J.R. et al. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat Cell Biol 21, 311–318 (2019). https://doi.org/10.1038/s41556-019-0282-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0282-9
This article is cited by
-
Interrogating epigenetic mechanisms with chemically customized chromatin
Nature Reviews Genetics (2024)
-
Decoding chromatin states by proteomic profiling of nucleosome readers
Nature (2024)
-
Transient chromatin compaction in fork restart
Nature Cell Biology (2023)
-
Targeting DNA damage response pathways in cancer
Nature Reviews Cancer (2023)
-
Genetic separation of Brca1 functions reveal mutation-dependent Polθ vulnerabilities
Nature Communications (2023)