Abstract
In cardiovascular tissues, changes in the mechanical properties of the extracellular matrix are associated with cellular de-differentiation and with subsequent functional declines. However, the underlying mechanoreceptive mechanisms are largely unclear. Here, by generating high-resolution, full-field strain maps of cardiomyocyte nuclei during contraction in vitro, complemented with evidence from tissues from patients with cardiomyopathy and from mice with reduced cardiac performance, we show that cardiomyocytes establish a distinct nuclear organization during maturation, characterized by the reorganization of H3K9me3-marked chromatin towards the nuclear border. Specifically, we show that intranuclear tension is spatially correlated with H3K9me3-marked chromatin, that reductions in nuclear deformation (through environmental stiffening or through the disruption of complexes of the linker of nucleoskeleton and cytoskeleton) abrogate chromatin reorganization and lead to the dissociation of H3K9me3-marked chromatin from the nuclear periphery, and that the suppression of H3K9 methylation induces chromatin reorganization and reduces the expression of cardiac developmental genes. Overall, our findings indicate that, by integrating environmental mechanical cues, the nuclei of cardiomyocytes guide and stabilize the fate of cells through the reorganization of epigenetically marked chromatin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, but they are available for research purposes from the corresponding author upon reasonable request. The RNA-seq data are available from the Gene Expression Omnibus under accession codes GSE109405 and GSE178674. Raw western blot data generated in this study are available from figshare with the identifier https://doi.org/10.6084/m9.figshare.16702888. Additional raw data underlying the bar graphs are available from figshare with the identifier https://doi.org/10.6084/m9.figshare.16702900. Source data are provided with this paper.
Code availability
Custom MATLAB codes to calculate chromatin marker intensity, track nuclear outlines, quantify the length of different sarcomere features, calculate marker co-localization scores and analyse nuclear strains are available from the corresponding author upon request.
References
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Phillip, J. M., Aifuwa, I., Walston, J. & Wirtz, D. The mechanobiology of aging. Annu. Rev. Biomed. Eng. 17, 113–141 (2015).
Lampi, M. C. & Reinhart-King, C. A. Targeting extracellular matrix stiffness to attenuate disease: from molecular mechanisms to clinical trials. Sci. Transl. Med. 10, eaao0475 (2018).
Thienpont, B. et al. The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy. J. Clin. Invest. 127, 335–348 (2017).
Zhang, Q. J. et al. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J. Clin. Invest. 121, 2447–2456 (2011).
Swift, J. et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013).
Fedorchak, G. R., Kaminski, A. & Lammerding, J. Cellular mechanosensing: getting to the nucleus of it all. Prog. Biophys. Mol. Biol. https://doi.org/10.1016/j.pbiomolbio.2014.06.009 (2014).
Heo, S.-J. et al. Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. Elife 5, e18207 (2016).
Uzer, G. et al. Cell mechanosensitivity to extremely low-magnitude signals is enabled by a LINCed nucleus. Stem Cells 33, 2063–2076 (2015).
Spagnol, S. T., Armiger, T. J. & Dahl, K. N. Mechanobiology of chromatin and the nuclear interior. Cell. Mol. Bioeng. 9, 268–276 (2016).
Crisp, M. et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172, 41–53 (2006).
Apel, E. D., Lewis, R. M., Grady, R. M. & Sanes, J. R. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275, 31986–31995 (2000).
Zhang, Q., Ragnauth, C., Greener, M. J., Shanahan, C. M. & Roberts, R. G. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics 80, 473–481 (2002).
Zhang, Q. et al. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 118, 673–687 (2005).
Guilluy, C. et al. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 16, 376–381 (2014).
Davidson, P. M. & Lammerding, J. Broken nuclei - lamins, nuclear mechanics, and disease. Trends Cell Biol. 24, 247–256 (2013).
Puckelwartz, M. J. et al. Nesprin-1 mutations in human and murine cardiomyopathy. J. Mol. Cell. Cardiol. 48, 600–608 (2010).
Banerjee, I. et al. Targeted ablation of nesprin 1 and nesprin 2 from murine myocardium results in cardiomyopathy, altered nuclear morphology and inhibition of the biomechanical gene response. PLoS Genet. 10, e1004114 (2014).
Hug, C. B. & Vaquerizas, J. M. The birth of the 3D genome during early embryonic development. Trends Genet. 34, 903–914 (2018).
Solovei, I. et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013).
de Las Heras, J. I. et al. Tissue specificity in the nuclear envelope supports its functional complexity. Nucleus 4, 460–477 (2014).
Joffe, B., Leonhardt, H. & Solovei, I. Differentiation and large scale spatial organization of the genome. Curr. Opin. Genet. Dev. 20, 562–569 (2010).
Parada, L. A., McQueen, P. G. & Misteli, T. Tissue-specific spatial organization of genomes. Genome Biol. 5, R44 (2004).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Pombo, A. & Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015).
Olins, A. L. & Olins, D. E. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 5, 30 (2004).
Kim, S. H. et al. Spatial genome organization during T-cell differentiation. Cytogenet. Genome Res. 105, 292–301 (2004).
Solovei, I. et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356–368 (2009).
Engler, A. J. et al. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell Biol. 166, 877–887 (2004).
Jacot, J. G., McCulloch, A. D. & Omens, J. H. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys. J. 95, 3479–3487 (2008).
Yahalom-Ronen, Y., Rajchman, D., Sarig, R., Geiger, B. & Tzahor, E. Reduced matrix rigidity promotes neonatal cardiomyocyte dedifferentiation, proliferation and clonal expansion. Elife 4, e07455 (2015).
Barnes, J. M., Przybyla, L. & Weaver, V. M. Tissue mechanics regulate brain development, homeostasis and disease. J. Cell Sci. 130, 71–82 (2017).
Porrello, E. R. et al. miR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 109, 670–679 (2011).
Young, J. L., Kretchmer, K., Ondeck, M. G., Zambon, A. C. & Engler, A. J. Mechanosensitive kinases regulate stiffness-induced cardiomyocyte maturation. Sci. Rep. 4, 6425 (2014).
Sreejit, P., Kumar, S. & Verma, R. S. An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell. Dev. Biol. Anim. 44, 45–50 (2008).
Terme, J.-M. et al. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J. Biol. Chem. 286, 35347–35357 (2011).
Barrero, M. J., Sese, B., Martí, M. & Izpisua Belmonte, J. C. Macro histone variants are critical for the differentiation of human pluripotent cells. J. Biol. Chem. 288, 16110–16116 (2013).
Xia, W. & Jiao, J. Histone variant H3.3 orchestrates neural stem cell differentiation in the developing brain. Cell Death Differ. 24, 1548–1563 (2017).
Valenzuela, N. et al. Cardiomyocyte-specific conditional knockout of the histone chaperone HIRA in mice results in hypertrophy, sarcolemmal damage and focal replacement fibrosis. Dis. Model. Mech. 9, 335–345 (2016).
Rose, B. A., Force, T. & Wang, Y. Mitogen-activated protein kinase signaling in the heart: angels versus demons in a heart-breaking tale. Physiol. Rev. 90, 1507–1546 (2010).
Riccio, A. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat. Neurosci. 13, 1330–1337 (2010).
Hogan, M. S., Parfitt, D. E., Zepeda-Mendoza, C. J., Shen, M. M. & Spector, D. L. Transient pairing of homologous Oct4 alleles accompanies the onset of embryonic stem cell differentiation. Cell Stem Cell 16, 275–288 (2015).
Filion, G. J. et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224 (2010).
Mysliwiec, M. R. et al. Jarid2 (Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J. Biol. Chem. 287, 1235–1241 (2012).
Dal-Pra, S., Hodgkinson, C. P., Mirotsou, M., Kirste, I. & Dzau, V. J. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by MIR combo. Circ. Res. 120, 1403–1413 (2017).
Bluemke, D. A. et al. The relationship of left ventricular mass and geometry to incident cardiovascular events. The MESA (Multi-Ethnic Study of Atherosclerosis) study. J. Am. Coll. Cardiol. 52, 2148–2155 (2008).
Brumback, L. C. et al. Body size adjustments for left ventricular mass by cardiovascular magnetic resonance and their impact on left ventricular hypertrophy classification. Int. J. Cardiovasc. Imaging 26, 459–468 (2010).
Akimoto, T. et al. Skeletal muscle adaptation in response to mechanical stress in p130cas−/− mice. Am. J. Physiol. Cell Physiol. 304, C541–C547 (2013).
Sawada, Y. et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015–1026 (2006).
Kovacic-Milivojević, B., Damsky, C. C., Gardner, D. G. & Ilić, D. Requirements for the localization of p130 Cas to Z-lines in cardiac myocytes. Cell. Mol. Biol. Lett. 7, 323–329 (2002).
Ni, Z. et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 28, 1161–1170 (2008).
Le, H. Q. et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18, 864–875 (2016).
Ghosh, S. et al. Deformation microscopy for dynamic intracellular and intranuclear mapping of mechanics with high spatiotemporal resolution. Cell Rep. 27, 1607–1620.e4 (2019).
Henderson, J. T., Shannon, G., Veress, A. I. & Neu, C. P. Direct measurement of intranuclear strain distributions and RNA synthesis in single cells embedded within native tissue. Biophys. J. 105, 2252–2261 (2013).
Lu, W. et al. Nesprin interchain associations control nuclear size. Cell. Mol. Life Sci. 69, 3493–3509 (2012).
Grady, R. M., Starr, D. A., Ackerman, G. L., Sanes, J. R. & Han, M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc. Natl Acad. Sci. USA 102, 4359–4364 (2005).
Iyer, K. V., Pulford, S., Mogilner, A. & Shivashankar, G. V. Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophys. J. 103, 1416–1428 (2012).
Auld, A. L. & Folker, E. S. Nucleus-dependent sarcomere assembly is mediated by the LINC complex. Mol. Biol. Cell 27, 2351–2359 (2016).
Balakrishnan, L. & Milavetz, B. Epigenetic regulation of viral biological processes. Viruses 9, 346 (2017).
Brumbaugh, J. et al. Inducible histone K-to-M mutations are dynamic tools to probe the physiological role of site-specific histone methylation in vitro and in vivo. Nat. Cell Biol. 21, 1449–1461 (2019).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013).
Herz, H. M. et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345, 1065–1070 (2014).
Sexton, T., Schober, H., Fraser, P. & Gasser, S. M. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 14, 1049–1055 (2007).
Honda, H. et al. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130(Cas). Nat. Genet. 19, 361–365 (1998).
Queisser, G., Wiegert, S. & Bading, H. Structural dynamics of the cell nucleus: basis for morphology modulation of nuclear calcium signaling and gene transcription. Nucleus 2, 98–104 (2011).
Stephens, A. D. et al. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 29, 220–233 (2018).
Alam, S. G. et al. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci. Rep. 6, 38063 (2016).
Spitzer, N. C. Electrical activity in early neuronal development. Nature 444, 707–712 (2006).
Wilkie, G. S. et al. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol. Cell. Proteomics 10, M110.003129 (2011).
Zuleger, N. et al. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol. 14, R14 (2013).
Mehta, I. S., Amira, M., Harvey, A. J. & Bridger, J. M. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 11, R5 (2010).
Kulashreshtha, M., Mehta, I. S., Kumar, P. & Rao, B. J. Chromosome territory relocation during DNA repair requires nuclear myosin 1 recruitment to chromatin mediated by ϒ-H2AX signaling. Nucleic Acids Res. 44, 8272–8291 (2016).
Aureille, J., Belaadi, N. & Guilluy, C. Mechanotransduction via the nuclear envelope: a distant reflection of the cell surface. Curr. Opin. Cell Biol. 44, 59–67 (2017).
Hoffman, L. M. et al. Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol. Biol. Cell 31, 1774–1787 (2020).
Pasqualini, F. S. et al. Traction force microscopy of engineered cardiac tissues. PLoS ONE 13, e0194706 (2018).
Xu, X., Li, Z., Cai, L., Calve, S. & Neu, C. P. Mapping the nonreciprocal micromechanics of individual cells and the surrounding matrix within living tissues. Sci. Rep. 6, 24272 (2016).
Worke, L. J. et al. Densification of type I collagen matrices as a model for cardiac fibrosis. Adv. Healthc. Mater. https://doi.org/10.1002/adhm.201700114 (2017).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–91 (2014).
Li, Q. et al. Selection of reference genes for normalization of quantitative polymerase chain reaction data in mouse models of heart failure. Mol. Med. Rep. 11, 393–399 (2015).
Martino, A. et al. Selection of reference genes for normalization of real-time PCR data in minipig heart failure model and evaluation of TNF-α mRNA expression. J. Biotechnol. 153, 92–99 (2011).
Pereyra, A. S. et al. Loss of cardiac carnitine palmitoyltransferase 2 results in rapamycin-resistant, acetylation-independent hypertrophy. J. Biol. Chem. 292, 18443–18456 (2017).
Chen, C. Y. et al. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat. Med. 24, 1225–1233 (2018).
Peyster, E. G. et al. In situ immune profiling of heart transplant biopsies improves diagnostic accuracy and rejection risk stratification. JACC Basic Transl. Sci. 5, 328–340 (2020).
Pugach, E. K., Blenck, C. L., Dragavon, J. M., Langer, S. J. & Leinwand, L. A. Estrogen receptor profiling and activity in cardiac myocytes. Mol. Cell. Endocrinol. 431, 62–70 (2016).
Chu, J. et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat. Methods 11, 572–578 (2014).
Acknowledgements
We thank L. Leinwand and S. J. Langer for providing materials and technical assistance for the generation of adenoviruses; N. Emery for biostatistical support and M. Rafuse for technical assistance with tissue sectioning; the Genomics Shared Resource Facility at the UC Anschutz Medical Campus and the Purdue Genomics Core Facilities for library preparation and RNA sequencing of our samples. This work was supported in part by grants to C.P.N. (NIH R01 AR063712, NIH R21 AR066230 and NSF CMMI CAREER 1349735) and to S.E.S. (NIH T32 GM065103).
Author information
Authors and Affiliations
Contributions
B.S., S.G. and C.P.N. conceptualized the study; B.S., S.G., A.K.S., C.J.G., S.C., J.B. and C.P.N. planned and implemented the methodology; software, B.S. and S.G.; formal analysis, B.S., S.G., S.E.S. and A.K.S.; investigation, B.S., A.K.S., S.E.S., A.G.B., K.B.M., K.B., E.C. and A.R.S.; B.S. wrote the original draft of the manuscript; all authors reviewed and edited the manuscript; C.P.N. acquired funding; Resources, C.J.G. and C.P.N.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Determining the ratio of contractile CMs to non-contractile cells in embryonic cardiac cultures on substrates with different stiffness.
a) Embryonic cardiac cells were isolated from (E)18.5 H2b-eGFP embryo hearts and cultured on soft (13 kPa) or stiff (140 kPa) PDMS. After two or four days, cultures were stained for actin and images of 300×300 µm2 areas were acquired. Using cell nuclei as reference, cells with clearly formed striated myofibrils were counted as contractile CM (*) or otherwise as non-contractile cells (+). b) Close-up shows the area indicated by a rectangle in the upper-left frame with adjusted intensity settings to accentuate myofibril striations.
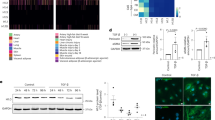
Extended Data Fig. 2 Stiff substrates reduce synchronization of calcium signaling, alter cell-ECM interaction pathways and inhibit stretch-activation of p130Cas in embryonic CMs.
a) Embryonic CMs (day E18.5) from H2b-eGFP mice were plated on either soft (13 kPa) or stiff (140 kPa) PDMS substrates for up to four days. At each day, a sample was stained with Fluo-4 AM to record Ca2+ activity as a proxy for CM contractility. b) Synchronicity was calculated as the average of the relative number of cells that beat together over a 20 second time window. c) Detailed analysis of histone family expression from RNAseq data of CMs plated on soft or stiff PDMS for four days (relative RPKM). d) Network analysis of global gene expression change revealed alterations in MAPK signaling and associated pathways that play a role in cell-substrate interaction. Rap1 signaling included the downregulation Bcar1 coding for p130Cas, a mechanosensitive protein located within the Z-disk lattice in CMs. e) Western blot analysis showed a reduction of stretch-induced tyrosine-410 phosphorylation of p130Cas on stiff PDMS compared to soft. SD; N = 3; Two-tailed T-test. * p < 0.05.
Extended Data Fig. 3 Contractile CMs and non-contractile cells show differences in the positioning of H3K9 trimethylated chromatin in vitro, and the effect of stiff mechanical environments on chromatin organization in non-contractile cells in vitro.
Embryonic cardiac cells were isolated from (E)18.5 H2b-eGFP embryo hearts and cultured on soft (13 kPa) PDMS substrates. a) Actin staining corresponding to the data from Fig. 3b to distinguish contractile CMs from non-contractile cells (NCC) via the formation of myofibrils; scales=5 µm. b) After two days in culture, contractile CMs (C) with clearly formed myofibrils showed enrichment of H3K9me3-marked chromatin at the nuclear border while actin-fiber forming NCCs (N) showed a more homogenous distribution throughout the nuclear interior. c) Cardiac cells were isolated from (E)18.5 H2b-eGFP embryo hearts and cultured on either soft (13 kPa) PDMS, stiff (140 kPa) PDMS or TCP for two or four days after which cells were stained for H3K27me3 and H3K9me3 as well as actin to distinguish non-contractile cells (NCCs) from CMs. Scales=5 µm. d) NCC nuclei were evaluated for peripheral enrichment (0 = center, 1=periphery) of overall chromatin (H2b) or epigenetically marked chromatin. Gray areas indicate center and peripheral bin; SEM; n ≥ 20 from 3 exp. e) Enrichment scores for each chromatin marker were calculated as the quotient of intensity of the peripheral bin (0.85-0.95) divided by the center bin (0.05-0.15). Substrate stiffness only minorly affected chromatin organization and enrichment of overall and H3K9me3-marked chromatin remained low while enrichment of H3K27me3-modified chromatin remained high throughout the four-day culture period. SEM; n ≥ 20 from 3 exp.; 1W-ANOVA: * p < 0.05, ** p < 0.01.
Extended Data Fig. 4 Histology and additional correlation data for patients with human cardiomyopathy.
a) Actin staining corresponding to the data from Fig. 4b for hypertrophic mice and Fig. 4g for human patients. Scales=5 µm. b) Masson’s trichrome staining of transmural cardiac tissue samples from human patients. Muscle tissue shown in red while collagen stains blue. Patients with nonischemic cardiomyopathy (NICM) show increased collagen deposition. Scales=100 µm c) Peripheral enrichment scores of DAPI and H3K27me3 were correlated with indicators of cardiac health. R2adj.=adjusted correlation coefficient.
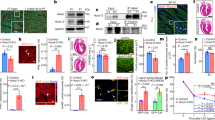
Extended Data Fig. 5 Stiff substrates reduce myofibril contractility in embryonic CMs.
a) CMs were infected with the adenoviral decoupling vector K3 (see Fig. 7a) or control vector CTL (shown) on day one and image series of myofibril contraction were recorded on day two via fluorescently tagged α-actinin 2. See also Supplementary Videos 1–3. Scale=10 µm. b) Top: α-actinin 2 intensity profile, as indicated by a red line in c), before (resting) and during contraction. Bottom: Close-up of two intensity peaks. Analysis of intensity profiles was used to determine the difference in length of overall sarcomeres (S), A-bands (A) and Z-disks (Z) during contraction. c) Control infected CMs on stiff PDMS (140 kPa, CTL) and decoupled CMs on soft PDMS (13 kPa, K3) showed inhibited contraction as overall sarcomere and A-band shortening as well as Z-disk extension was abrogated compared to control infected CMs on soft PDMS (13 kPa, CTL); n = 25 from 5 exp.; 1W-ANOVA. d) Image series of CM nuclei cultured on either soft (13 kPa) PDMS, stiff (140 kPa) PDMS or TCP were recorded during contraction and bulk linear strain and translational movement of nuclei were determined over four days. Nuclei of CMs cultured on soft substrates showed higher bulk linear strain and translational movement compared to stiff PDMS and TCP; SEM; n > 44 from 4 exp.; 1W-ANOVA. e) Bulk linear strain and translational movement of CM nuclei were determined after LINC disruption on day two (24 h post infection) and day four. Decoupled nuclei (K3, n = 32) showed lower bulk linear strain and translational movement compared to cells infected with the control vector (CTL, n = 32) or non-infected control cells (NIC, n = 67); SEM; from 4 exp.; 1W-ANOVA. f) Image stacks of beating CM nuclei were recorded on day two after which cells were stained for H3K9me3 and H3K27me3. Analysis of the translational movement of the nucleus and of dense, H3K9me3-rich heterochromatin clusters showed, that movement was higher for intra-nuclear heterochromatin than bulk movement of their respective nuclei during contractions. White and red outlines indicate nuclear and cluster boundary during rest and peak contraction, respectively. n = 10 from 3 exp.; T-test; scale=5 µm. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Extended Data Fig. 6 Colocalization of chromatin markers with myofibrils in CMs and emerin localization in the outer nuclear membrane.
a) Embryonic cardiac cells from day (E)18.5 H2b-eGFP mice were cultured for four days after which they were stained for different markers and z-stacks were recorded on a confocal microscope. Left: Z-projection as well as XZ and YZ slices along white dashed lines for a CTL infected CM. Right: Panels show representative z-slices at different z-positions (basal, medial, apical) indicated by white arrows in the XZ projection. Basal z-slices were used for marker overlap analysis. b) After four days in culture soft (13 kPa) or stiff (140 kPa) PDMS or TCP, CMs were stained for emerin using digitonin to selectively permeabilize the cell membrane but not the nuclear membrane. Emerin localization at the outer nuclear membrane was similar for all substrates; scales=5 µm.
Extended Data Fig. 7 Extended analysis of intranuclear strains during CM contraction.
Embryonic CMs were cultured on soft (13 kPa) PDMS for two days. Intranuclear strain maps of CM nuclei during contraction were generated via deformation microscopy after which cells were stained for H3K9me3, H3K27me3 or actively transcribed chromatin (RPIIS2) and strain occupancy for chromatin markers was analysed (see Fig. 6). a) Intranuclear strains were analysed over chromatin marker intensities. b, c) Intranuclear strains and chromatin marker intensities were analysed independent of each other with respect to chromatin density as judged by H2b intensity. Chromatin density distribution (histogram) is represented as relative count on the right y-axis. Hydrostatic strains are lowest around medium chromatin density (density histogram peak) and increases for denser chromatin which is primarily occupied by H3K9me3 modifications. SEM; n = 20 from 5 exp. d) Intranuclear strains were analysed over distance to the nuclear center. Strains declined towards the nuclear border, excluding the possibility of increased strain association of H3K9me3-marked chromatin due to its proximity to the periphery. e) Visual representation of different strain types. f) Embryonic CMs were cultured on soft (13 kPa) PDMS for two days after which image stacks of CM nuclei were recorded during contractions to determine the direction of nuclear translation. Cells were then stained for chromatin markers H3K9me3, H3K27me3 or actively transcribed chromatin (RPIIS2). Chromatin marker occupancy was calculated with respect to the angle of the nuclear center with the angle of nuclear translation set to 0°. Cells with extended major axis during contraction (tensile loading mode, n = 20, same as intranuclear analysis) showed a distinct peak of H3K9me3 intensity ±30° around the direction of translation while a decline in H3K9me3 intensity was observed for cells with shortened major axis (compressive loading mode, n = 8). Right side provides a graphic illustration of angular analysis showing nuclear outlines during resting phase (doted black) and peak contraction (solid green). The black arrow indicates the direction of translation, which defines the 0° point, and green arrows demonstrate extension or compression of the nuclear major axis used to determine the loading mode of cells. Error=SEM; from 5 exp.
Extended Data Fig. 8 LINC complex disruption in CMs.
a) On day one of culture (24 h after seeding), CMs were infected with an adenoviral vector that disrupted LINC connections (K3) or a control vector (CTL). 24 h post infection, CMs were fixed and stained for nesprin-1 after which widefield images were acquired. The decoupling vector showed successful integration of the truncated nesprin construct (mNep2.5) into the outer nuclear membrane while no distinct localization was observed for the control vector. Decoupled cells showed disrupted myofibril formation, particularly around the nucleus, and diminished presence of nesprin-1 at the nuclear membrane. Scales=5 µm. b) Images of infected cells plated on either soft (13 kPa) or stiff (140 kPa) PDMS. Decoupled cells show disrupted sarcomere fibers, particularly around the nucleus. Cells correspond to Extended Videos 1–3. Scales=10 µm. c) Embryonic cardiac cells were isolated and cultured on soft (13 kPa) PDMS. CMs were infected at day 1 and stained for actin and H3K27me3 on day two (shown) and day four. d) Decoupled cells (K3) showed abolished enrichment of overall and H3K9me3-marked chromatin compared to infected control cells (CTL) while H3K27me3-marked chromatin was similarly enriched (see also Fig. 7c). Scales=5 µm.
Extended Data Fig. 9 Colocalization of chromatin markers after LINC disruption.
a) Embryonic cardiac cells were isolated and cultured on soft (13 kPa) PDMS. CMs were infected with the LINC decoupling vector (K3) or control construct (CTL) on day one and stained for H3K9me3 or H3K27me3 on day four to analyze marker overlap (see Fig. 7). b) Colocalization of H3K9me3 and H3K27me3 with overall chromatin did not change after LINC disruption.
Extended Data Fig. 10 Inducible expression of H3.3 K-to-M mutant inhibits H3K9 trimethylation and changes cardiac gene expression, and H3K9me3 effects gene expression globally, but not locally.
a) Embryonic cardiac cells from H3K9M and H3WT mice were isolated and cultured on soft (13 kPa) PDMS. CMs were induced using 2 µg/ml doxycycline, harvested on day four, and subjected to Western blot analysis for H3K9me3 or H3 as a loading control. b) Functional annotation for top genes differentially regulated between H3K9M and H3WT cells, which show a phenotype consistent with CM development. c) RNAseq data of CMs from H3K9M and H3WT was analyzed with respect to their distance to H3K9me3 heterochromatin domains, using a ChIPseq dataset of 16.5-day mouse embryo hearts from modEncode. Left: The combined expression of H3K9M and H3WT showed a general decrease of gene expression with proximity to heterochromatin domains. Right: Relative expression in H3K9me3 suppressed H3K9M mice, compared to H3WT, showed no direction of expression change, only higher variance. d) Example of a single gene track for Gata-4. Repressive polycomb domains are shown in purple (ReprPC). Although there is a repressive polycomb domain near the TSS of this gene of interest there is no significant upregulation of the level of gene expression according to our RNA-seq data.
Supplementary information
Supplementary Information
Supplementary tables.
Supplementary Video 1
CMs from H2b-eGFP mice were plated on 13 kPa PDMS substrates and infected with the adenoviral control vector CTL (Fig. 6a) on day 1. Image series of myofibril contractions were recorded on day 2 via fluorescently tagged α-actinin 2 using an inverted wide-field microscope.
Supplementary Video 2
CMs from H2b-eGPP mice were plated on 140 kPa PDMS substrates and infected with the adenoviral control vector CTL (Fig. 6a) on day 1. Image series of myofibril contractions were recorded on day 2 via fluorescently tagged α-actinin 2 using an inverted wide-field microscope.
Supplementary Video 3
CMs from H2b-eGFP mice plated on 13 kPa PDMS substrates and infected with the adenoviral decoupling vector K3 (Fig. 6a) on day 1. Image series of myofibril contractions were recorded on day 2 via fluorescently tagged α-actinin 2 using an inverted wide-field microscope.
Source data
Source Data Fig. 3
Source data.
Source Data Fig. 4
Source data.
Source Data Fig. 7
Source data.
Source Data Fig. 8
Source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Source data.
Source Data Extended Data Fig. 5
Source data.
Rights and permissions
About this article
Cite this article
Seelbinder, B., Ghosh, S., Schneider, S.E. et al. Nuclear deformation guides chromatin reorganization in cardiac development and disease. Nat Biomed Eng 5, 1500–1516 (2021). https://doi.org/10.1038/s41551-021-00823-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00823-9
This article is cited by
-
Dysregulated CREB3 cleavage at the nuclear membrane induces karyoptosis-mediated cell death
Experimental & Molecular Medicine (2024)
-
Cardiovascular Mechano-Epigenetics: Force-Dependent Regulation of Histone Modifications and Gene Regulation
Cardiovascular Drugs and Therapy (2024)
-
Chromatin organization and behavior in HRAS-transformed mouse fibroblasts
Chromosoma (2024)
-
Distinguishable DNA methylation defines a cardiac-specific epigenetic clock
Clinical Epigenetics (2023)
-
InterLINCing Chromatin Organization and Mechanobiology in Laminopathies
Current Cardiology Reports (2023)