Abstract
Membrane biofouling poses an ongoing challenge in seawater reverse osmosis (SWRO) desalination. Here we delved into the impact of seasonal variations in microbial communities on membrane biofouling and innovatively fabricated quorum sensing inhibitors (acylase (AC) and methyl anthranilate (MA))-modified membranes to combat it. Results indicated that Proteobacteria dominated in seawater and membrane biofilm across all seasons, while other phyla varied seasonally. At the class level, the two leading bacteria on the membrane were Gamma- and Alphaproteobacteria, constituting 14–48% and 4–27%, respectively. Genera like Arcobacter, Vibrio, and Rhodobacteraceae were identified as keystone species that exhibited a significant positive correlation with extracellular polymeric substance (EPS) and biofilm formation, leading to a substantive reduction in membrane flux by 70 to 77%. The introduction of AC and MA inhibitors on the membrane suppressed keystone bacteria Rhodobacteraceae and Arcobacter and affected their metabolism, thereby significantly reducing EPS by 65–69% and 55–59%, respectively, and alleviating membrane flux decline by 30–32% and 18–22%, respectively, compared to the pristine membrane. These findings shed new light on the seasonal patterns of membrane biofouling and provide valuable insights into anti-biofouling strategies based on QS inhibition for collaborative biofilm formation.
Similar content being viewed by others
Introduction
As a rich water resource, seawater is considered an excellent alternative to solve freshwater scarcity. Reverse osmosis (RO) desalination has been widely applied (around 69% of the installed capacity globally)1,2, resulting in high-quality desalted water and low operation costs3. Nevertheless, membrane biofouling occurred due to the attachment and proliferation of bacteria on the membrane surface, forming a biofilm containing secreted extracellular polymeric substances (EPS)4. It has been referred to as the critical weakness of RO membrane technology, causing issues for the majority (~70%) of the SWRO systems5. Chlorine-based chemicals are commonly utilized for pretreatment; however, the free chlorines (e.g., hypochlorous) can damage the active layer of the RO membrane6. Some biocides, such as tributyl tetradecyl phosphonium chloride7 and ethyl lauroyl arginate8, can induce the emergence of resistant bacteria. Moreover, the dead bacteria serve as source of nutrients for living cells, exacerbating biofouling. Only less than 0.01% of the surviving bacteria are sufficient to form another mature biofilm9. An alternative without adverse effects for controlling biofouling needs yet to be proposed.
Quorum sensing (QS) is a bacterial communication process that activates the expression of related genes through signaling molecules to coordinated behaviors such as bacterial virulence, bioluminescence, motility and biofilm formation10. The QS inhibitors (QSI) can thus disrupt bacterial communication pathways and offer advantages of eco-friendliness, high biofilm inhibitory activities, and low toxicity10, which show a non-disinfectant biological alternative approach to mitigate membrane biofouling. Several studies have employed inhibitors to alleviate membrane biofouling. Recent endeavors have demonstrated that the biofilm formation on the membrane surface by P. aeruginosa was reduced through acylase (AC) and methyl anthranilate (MA) based on the inhibition of Acyl-homoserine lactones (AHL) and Pseudomonas quinolone signal (PQS) signaling pathways11,12. The downside of using directly dispersed inhibitors is the likelihood of losing their inhibition efficiency and the secondary pollution caused by inhibitors. It has been confirmed that enzymatic activity was increased by 90% after immobilizing the acylase enzyme onto a nanofiltration membrane compared to the free enzyme13. Therefore, membrane modification incorporating QSI is a promising approach to improve membrane performance.
Thus far, membrane biofouling has received widespread attention in the membrane treatment process. Numerous laboratory studies have explored the formation process of biofouling. However, most of these studies were performed with model bacteria, and due to the diversity of microorganisms in seawater sources, microbial species rarely exist alone, which makes it challenging to represent real biofouling in practical applications. In particular, significant differences in the microbial communities on the RO membrane biofilm during the winter, summer, and autumn seasons have been observed14,15. Thus, seasonal variations in the composition and concentration of EPS secreted by bacteria16,17 may lead to different fouling behaviors in SWRO. These results show that the effects of microbial community changes according to seasons are significant to the dynamics of biofilm formation and membrane fouling control.
Our previous studies have demonstrated that AC and MA alleviated the membrane flux reduction by reducing the EPS secretion and biofilm formation in RO and FO filtration process11,12. Besides, the introduction of MA on the PVDF membrane suppressed the expression of PQS and AHL synthesis genes in Pseudomonas aeruginosa, and thus mitigated biofilm formation on the modified membrane9. However, the roles of QSIs in seawater microbial interspecies communication and community assemblages have yet to be experimentally investigated or theoretically addressed. Therefore, this study investigated the response of seawater microbial communities and biofilm microbiota to seasonal changes and the roles of QSIs in regulating microbial communities. The effects of microbial diversity on membrane biofouling were then evaluated. The main objectives of the study include: (1) investigating the impact of seasonal changes on bacterial community succession in the SWRO membrane desalination process; (2) revealing the correlation between seasonal bacterial community and SWRO membrane biofouling propensity, and clarifying the key bacterial groups based on phylogenetic molecular ecological networks (pMENs) and heatmap factor analysis, and (3) examining the effects of AC- and MA-modified membranes on seasonal fouling behaviors and their control.
Results and discussion
Characteristics of QSI-modified membranes
Anti-biofouling SWRO membranes were prepared by incorporating inhibitors, i.e., AC and MA, via Schiff base reaction. The physicochemical and microscopic characterizations verified the successful incorporation of AC and MA on the membrane (Supplementary Fig. 1). Specifically, the peak at 1710 cm−1 represents the stretching vibration of C = O18 that originates from the GA load. After loading the inhibitors, however, the stretching vibration of C = O disappeared, and the peak at 1671 cm−1 ascribed to the C = N stretching19 has increased on the AC_m and MA_m membranes (Supplementary Fig. 1a), suggesting a reaction between the aldehyde group of GA and the end amino groups of AC and MA by Schiff base reaction20. The N 1s spectra indicate that the peaks at a binding energy of 399.8 and 401.7 eV correspond to the N atoms in amine and amide moieties21, respectively. The increase in the C-N peak area is attributed to the formation of AC and MA layers (Supplementary Table 1).
The pristine membrane exhibited a negative charge at pH = 5.5–10 owing to the carboxylic acid functional group of the polyamide layer22 (Supplementary Fig. 1d). After coating the PDA layer, the surface charge increased due to the free amine and other nitrogen-containing groups23. The surface charges of the GA_m, AC_m and MA_m membranes were lower than that of the PDA_m membrane because of the conversion of the amino group to the C=N structure20. The hydrophilicity of the modified membranes was determined by the contact angle. The water contact angles of the pristine membrane, AC_m, and MA_m membranes were 45, 40, and 32, respectively (Supplementary Fig. 1e). AC_m and MA_m membranes possessed more hydrophilic properties than the pristine membrane owing to the introduction of hydrophilic phenolic hydroxyl, amino groups and pendant-CHO by PDA and GA24,25. The 10-min decline rate of the contact angle of AC_m and MA_m membranes (decreased 65% to 14 and 76% to 7.5) was more significant than that of the pristine membrane (reduced 50% to 22), suggesting the better wettability of the AC_m and MA_m membranes. More details for the surface morphology characteristics of QSI-modified membranes are shown in Supplementary Fig. 1.
Dynamics of SWRO biofouling with seasonal variation
Supplementary Table 2 show physical-chemical and biological fluctuations for four seasons (i.e., January—winter, May—spring, July—summer, and October—fall) of natural seawater (NSW) collected from the Bohai Bay (38.97N, 117.77E; Tianjin, China) during this study period. The conductivity peaked in the winter and was negatively correlated with water temperature. The total bacterial cells reached a peak in the summer, coinciding with a previous finding26. The pH values of feedwater for all experiments varied between 7.8 and 8.2, and the dissolved organic carbon (DOC) level was maintained between 0.66 and 1.43 mg L−1.
We conducted dynamic biofouling experiments on SWRO membranes using seawater according to seasons and compared the results with those of QSI-modified membranes in a crossflow system. The normalized membrane flux changes, as depicted in Fig. 1a, reveal that the pristine SWRO membrane experienced the most substantial flux decline, with a total flux decreasing from 70 to 77%. This confirms the severe fouling propensity by seawater, with the most pronounced water flux decline observed in the summer (77%).
The changes in a normalized permeate flux and b salt rejection during dynamic biofouling with seasonal variations in the filtration of control (WI_control, SP_control, SU_control, and FA_control), AC_m (WI_AC_m, SP_AC_m, SU_AC_m, FA_AC_m), and MA_m (WI_MA_m, SP_MA_m, SU_MA_m, and FA_MA_m) membranes.
In contrast, the flux decline of AC_m and MA_m membrane was alleviated by 30–32% and 18–22%, respectively, suggesting that modification with QSI effectively mitigated membrane fouling under dynamic crossflow filtration conditions, although their extent still depended on seasons. Furthermore, the AC_m and MA_m membranes maintained slightly but greater salt rejection rate (98.5%) compared to the pristine membrane (97.5%), which can be ascribed to the lower flux decline (i.e., fouling) of QSI-modified membranes than the pristine RO membrane.
Seasonal variation in EPS composition and biofilm structure
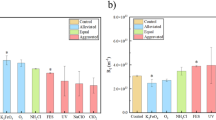
To further characterize the biofouling behaviors of pristine and QSI-modified membranes, the EPS (i.e., proteins, polysaccharides, and eDNA), TOC, and biofilm were assessed under different seasonal conditions. The EPS content on the pristine membrane was found to be higher during the summer compared to the other three seasons in SWRO. Specifically, the EPS content values were 104 μg cm−² for WI_control, 110 μg cm−² for SP_control, 129 μg cm−² for SU_control, and 118 μg cm−² for FA_control (Fig. 2a). This observation is consistent with the result of the higher contents of EPS extracted from the microbial biofilm in the summer27. The mass ratio of polysaccharides to proteins is associated with biofilm strength28. The greater the ratio of polysaccharides to proteins, the higher the level of attachment within the biofilm. It is essential to highlight that the relative polysaccharide content in SP_control, SU_control, and FA_control was 44, 42, and 43%, respectively. This was significantly higher than WI_control, which had a content of 23%. These findings suggest notable distinctions in biofouling formation during the winter season.
a polysaccharides, proteins, TOC and b eDNA concentrations. c The total cell fluorescence (live and dead cells), live/total cell ratio on the fouling layer. d Biofilm thickness was analyzed by confocal laser scanning microscopy. Asterisks (*) indicate a statistically significant difference between the control and QSI-modified membranes (*P < 0.05). Error bars represent one standard deviation of triplicate experiments.
Polysaccharides and proteins are major EPS components, among which eDNA was also reported as a “glue” contributing to biofilm stability even at low abundance29. As shown in Fig. 2a, the AC_m and MA_m membranes significantly decreased the EPS contents by 65–69% and 55–59%, respectively, compared to the control membrane. The contents of polysaccharides, proteins, and eDNA (Fig. 2b) decreased on the AC_m membrane surface by 42–62%, 72–75%, and 59–65%, respectively, while on the MA_m membrane, they decreased by 47–49%, 60–66%, and 50–57%, respectively. The results revealed that QSI-modified membranes reduced EPS production by marine consortial bacteria, especially protein secretion. The lower organic biometabolic components on the QSI-modified membranes might be due to the fact that inhibitors can interfere with the release of EPS (i.e., proteins, polysaccharides, and eDNA). Hence, the fluctuations in EPS could substantiate the patterns observed in the flux behavior depicted in Fig. 1a.
The CLSM analysis revealed the biofilm structure and microbial activity on the membrane. In the control groups, the biofilm completely covered the membrane surface, forming a thick and dense structure with a homogeneous mat (Supplementary Fig. 2). The result is consistent with a previous report15. In contrast, the biofilm formed on the modified membranes was very thin. As shown in Fig. 2c, d, the AC_m and MA_m membranes significantly reduced the total cell number by 70–73% and 60–67% while decreasing the average biofilm thickness from 60 to 17.5 μm and 20 μm, respectively, compared to the pristine membrane. Meanwhile, the AC_m and MA_m membranes exhibited a similar ratio of live/total cells as the pristine membrane. The results indicate that the AC_m and MA_m membranes delay bacterial growth without bactericidal properties, which resulted in slower biofilm formation and lesser EPS secretion.
Because the characteristics and content of biofilm/EPS are closely related to the microbial community composition30,31, its further analysis was conducted alongside metabolisms during SWRO.
Alpha and beta diversities
To characterize the microbial communities in SWRO membrane biofouling, we conducted 16S rRNA gene high-throughput sequencing. Each group demonstrated a high coverage index (>0.998), ensuring high credibility and accurate representation of the microbial community30. (Supplementary Table. 3). The Shannon values in seawater were in the order of FA (5.470505) > SU (4.668346) > SP (3.563673) > WI (3.146628) (Fig. 3a), suggesting that the diversity of microbial communities in seawater is greater during the warm season compared to colder seasons. A previous study on microbial diversity changes support the findings of this study16. As shown in Fig. 3b, c, the richness and diversity of biofilms formed on the membrane significantly decreased (P < 0.05), indicating that specific microorganisms in seawater can cause membrane biofouling. Among them, the microbial diversity of biofilm formed in winter was the lowest (Fig. 3a). The differences in the microbial community were also supported by the beta diversity analysis shown in Supplementary Fig. 3. Additionally, the AC_m and MA_m membranes reduced the Shannon index compared to the pristine membrane (Fig. 3b), indicating a decrease in the diversity of the biofilm developed on the QSI-modified membrane.
Seasonal dynamics of microbial communities in seawater and membrane biofilm
The variation of the microbial communities had a tremendous impact on the membrane biofouling32. We further explored the changes in taxa from the phylum to the genus level and demonstrated key species for the system performance. In seawater, taxonomic classification revealed that Proteobacteria predominated in all seawater samples throughout the whole experiment ranging from 58 to 64%, followed by Bacteroidetes (11 to 32%) (Fig. 4a), in which Alphaproteobacteria and Gammaproteobacteria were the dominant classes ranging from 48 to 59%. Seawater in the summer and fall seasons contains more Proteobacterial classes (Alpha-, Gamma-, Beta-, Delta-, and Epsilon-proteobacteria) than in winter and spring. It had been reported that Cyanobacteria showed significant enrichment under 5 °C compared to the samples at 15 and 20 °C33. In this study, Cyanobacteria displayed a higher prevalence, constituting 24% of the population during the winter season, whereas they were virtually absent during the spring, summer, and fall. This observation suggests their enhanced adaptability to cold-weather environments.
WI, SP, SU, and FA represent winter, spring, summer, and fall, respectively. a Phyla (which represent >3% of sequences) and the Proteobacterial class (inset) distributions are presented as the percent of total communities for the seawater and control membrane groups; b Linear discriminant analysis (LDA) effect size (LEfSe) analysis of microbial abundance with cladogram of microbial communities in comparison of seawater and control membrane; c Heatmap of the genus level composition of the top 20 species.
In the membrane biofilm, the dominant phyla were Proteobacteria (44–59%), followed by Bacteroidetes (21–47%) (Fig. 4a). This is consistent with previous research on microbial communities in full-scale desalination plants1,34,35. Nevertheless, the proportions of various Proteobacterial classes exhibited significant disparities between in seawater and on the membrane based on the LEfSe analysis (Fig. 5b). Alpha- (4–27%) and Gammaproteobacteria (14–48%) were dominant in the biofilm, while Epsilonproteobacteria had a low abundance (3–8%) and Delta- and Betaproteobacteria were nearly absent. Alphaproteobacteria was dominated in the seawater, yet its relative abundance varied significantly for the different biofilm samples. It was also observed that the biofilms developed on the SWRO membrane during four different seasons had different community structures, and the dominant bacteria varied with seasons. This is possibly because the seasonal seawater variations rendered a selective pressure on the bacteria responsible for membrane biofouling34. Specifically, although Alphaproteobacteria are the dominant seawater bacteria in winter and spring, the biofilm microbiota on the membrane shifted to Gammaproteobacteria dominance (Fig. 4a). This phenomenon is likely because, in winter and spring, the Alphaproteobacteria microbiota consisted of the genera Sulfitobacter, Lentibacter, Loktanella, and Planktomarina, which are not easily enriched on the membrane, as indicated in Fig. 4c. At the genus level in biofilm microbiota (Fig. 4c), the Lutibacter, Shewanella, and Vibrio belonging to the Gammaproteobacteria, and Flavobacterium belonging to the Bacteroidetes predominated the biofilm microbiota on the membrane surface in winter. Compared to winter, Rhodobacteraceae and Erythrobacter belonging to the Alphaproteobacteria, Acinetobacter belonging to Gammaproteobacteria, and Tenacibaculum belonging to the Bacteroidetes, have higher abundance in the membrane biofilm microbiota in spring, summer, and autumn. In addition, Arcobacter, which belongs to Epsilonproteobacteria, had a high abundance on the membrane in all four seasons.
a Z-P plot of OTUs in biofilm microbiomes based on pMENs and b Heatmap analysis for the correlation between the bacteria and fouling factors. Note: The X and Y axes are fouling factors and bacteria, respectively. R is shown with different colors, and the right side of the legend displays the color range of different R values. Significance is represented as: (*) for 0.01 < P < 0.05 and (**) for 0.001 < P < 0.01.
pMENs to identify keystone populations and correlation analysis
Based on operational taxonomic units (OTUs) results at the normalized sequencing depth (Supplementary Fig. 3c), the Venn diagram revealed that 59 OTUs were the dominant bacteria causing biofouling on the SWRO membrane. At the genus level, Vibrio, Rhodobacteraceae, Flavobacterium, Shewanella, Lutibacter, Arcobacter, Acinetobacter, Tenacibaculum, and Erythrobacter were the dominant genera in biofilm microbiota on the membrane (Fig. 4c). As reported previously, certain members of Alphaproteobacteria (e.g., Rhodobacteraceae and Erythrobacter) were identified as the primary colonial strain15,36,37; Vibrio (Gammaproteobacteria) is a well-known marine bacterium with high potential of biofilm formation38; Arcobacter (Epsilonproteobacteria) plays a vital role in the later development and maturation of biofilms39. The characteristics of the microbial community are related to the metabolic secretion of EPS40,41. The relationships between bacteria and biopolymers would be a significant indicator for the fouling analysis. However, few studies focus on the interaction between bacteria and biopolymers. In our study, a strong taxa-biopolymer relationship was observed in pMENs, as revealed by the Spearman correlation test (shown in Supplementary Fig. 4 and Fig. 5a, b). It was found that the keystone taxa Rhodobacteraceae, Vibrio and Arcobacter had a strong positive relationship (P ≤ 0.01) with EPS and biofilm contents. Alphaproteobacteria (Rhodobacteraceae) and Acinetobacter were positively correlated (P ≤ 0.01) with the polysaccharides (Fig. 5b), and their abundance on the SP_, SU_ and FA_control membranes was higher than that on the WI_control membrane (Fig. 4c). Therefore, the secretion of polysaccharides by these bacteria is likely to account for the relatively higher concentration of polysaccharides in EPS detected on the membrane in the spring, summer and fall seasons compared to winter (Fig. 2a). Not only did the species (Rhodobacteraceae, Vibrio, Acinetobacter, and Cryomorphaceae) enrich on the SU_control membranes (Fig. 4c), but they also exhibited a positive relationship with EPS and biofilm content (Fig. 5b). Additionally, the heatmap analyses for the correlation between the bacteria and fouling factors depending on seasons are shown in Supplementary Fig. 5. In summer, Erythrobacter (Alphaproteobacteria-affiliated bacteria) showed a strongly positively correlation with the EPS and biofilm contents. These findings may corroborate the most severe biofouling on the membrane in summer (Fig. 1). Furthermore, the non-keystone taxa Acinetobacter, Cryomorphaceae, and NS7_marine-group also had a high positive relationship (P ≤ 0.05) with polysaccharides. These species may be the potential contributors to membrane biofouling. Hence, their strong correlation with fouling factors is compelling evidence supporting their significant role in causing membrane fouling.
Biofilm microbiota responding to QS inhibitions
Changes in microbial composition on the QSI-modified membranes compared to the control membrane could reflect the effect of inhibitors on biofilm microbiota (Supplementary Fig. 6 and Fig. 6a). To explore the classified bacterial taxa with significant differences in abundance between the biofilm microbiota on the AC_m/MA_m membranes and the control, we performed biomarker analysis using the linear discriminant analysis (LDA) effect size (LEfSe) method42 (Fig. 6b, d). As shown in the LEfSe bar chart (Supplementary Fig. 7), 14 and 11 biomarkers with high LDA values (over 3.5) showed significant differences in the AC_m and MA_m membranes. Specifically, at the phylum level, both AC_m and MA_m significantly (P < 0.05) decreased the relative density of Proteobacteria from 51.1 to 21.8% and 29.7% (Fig. 6c, e), respectively compared to the control. However, bacteria responded differently to AC and MA, and AC has a more obvious inhibition effect on Proteobacteria, showing that the effects of AC and MA on some bacteria are different.
WI, SP, SU, and FA represent winter, spring, summer, and fall, respectively. a Phyla (which represent >3% of sequences) and the Proteobacterial class (inset) distributions are presented as the percent of total communities for the seawater and control membrane groups; Linear discriminant analysis (LDA) effect size (LEfSe) analysis of microbial abundance with cladogram of microbial communities in comparison of control and QSI-modified membranes (b AC_m and d MA_m); Relative abundances of the phyla, classes, genera that showed significant differences between control and QSI-modified membranes (c AC_m and e MA_m). A Kruskal‒Wallis method was used to evaluate the significance of differences between the indicated groups. *P < 0.05.
The introduction of AC and MA on the SWRO membrane possibly regulates the EPS-favorable microorganisms. The heatmap analysis showed that Gammaproteobacteria and Epsilonproteobacteria were positively correlated with EPS and biofilm (Supplementary Fig. 4b). Epsilonproteobacteria has been reported to be a critical bacterium in biofilm formation, and many species in this class contain the LuxS/Autoinducer-2 QS system43. The AC_m significantly (P < 0.05) reduced the relative abundance of Gammaproteobacteria by 72%, and the MA_m membrane decreased the relative abundance of Epsilonproteobacteria by 90%. At the family and genus level, Rhodobacteraceae and Arcobacter are frequently detected in biofilm, with the capacity of AHL-producing and EPS-secreting functions39,44,45. In our study, Rhodobacteraceae, Vibrio, and Arcobacter were perceived as contributors to EPS secretion and biofilm formation (as discussed in Fig. 5). The MA_m membrane significantly reduced the relative abundance of Arcobacter by more than 90%, and Rhodobacteraceae was nearly absent on the AC_m membrane (p > 0.05) (Fig. 6c, e). Furthermore, AC_m and MA_m membranes enhanced the abundance of quorum quenching (QQ)-related genera in biofilm microbiota (shown in Supplementary Fig. 8), such as Flavobacterium46, marine bacterium Tenacibaculum47 and Glutamicibacter (possessing AHL degradation activity)48, which were found to display efficient activities against biofilm formation and were prone to control membrane fouling49. These results indicated that the AC and MA introduction to the SWRO system would inhibit the proliferation of EPS-related bacteria and reduce their contribution to membrane biofouling. Therefore, this study indicated that investigating Proteobacteria-affiliated bacteria in the context of membrane biofouling under QS inhibition holds significant promise.
Effects of QSI-modified membranes on metabolisms in biofilm microbiota
Metabolic analysis was performed to elucidate the role of QS inhibitors on the metabolic activity of the bacteria on the membrane. The functions of KEGG were explored to obtain more information on bacterial metabolism. Amino acids and carbohydrate metabolism could be involved in the glycine/serine/threonine and amino/nucleotide sugar metabolism pathways (map00260 and map00520), as well as in the synthesis of extracellular proteins and polysaccharides50. QS ensures the successful establishment and survival of biofilm communities by coordinating attachment, matrix production, maturation, and resistance mechanisms51. In this study, we found that the functional abundance of amino acids and carbohydrate metabolism, QS, and biofilm formation was lower in the AC_m and MA_m membranes group than those in the control (Fig. 7), thus effectively mitigating biofouling formation.
Specifically, the relative metabolites and metabolism pathways based on the bacterial genome are depicted in Fig. 8, and typical genes involved in carbohydrate/amino acid metabolisms and their abundance are given in the Supplementary Table. 4. A group of sugar metabolites, such as glucose, galactose, and UDP-N-acetylglucosamine, were detected, which are involved in amino/nucleotide sugar metabolism and play a critical role in polysaccharide synthesis50. The critical genetic expressions (e.g., glmU and galU/galF) of glucose and N-acetylglucosamine were significantly reduced in the AC_m and MA_m membranes. Glycine, serine, and threonine metabolism are involved in regulating various pathways of valine, leucine, and isoleucine biosynthesis. Among them, valine, leucine, and isoleucine are essential metabolites for synthesizing many secondary metabolites52 and participating in protein synthesis. The abundance of genes serB and ilvE that regulate the generation of serine, valine, leucine, and isoleucine was reduced by 38 and 35% in the AC_m membrane, and decreased by 36 and 33% in the MA_m membrane, respectively, when compared with the control. Additionally, the abundance of genes (hom) encoding for the homoserine was significantly decreased in the MA_m membrane, and the abundance of encoding genes (ilvD) responsible for the oxopentanoic acid and 2-oxoisovalerate was significantly decreased in the AC_m membrane compared to the control.
Metabolic pathways enriched for significantly altered metabolites in the QSI-modified membranes (AC_m and MA_m) samples compared to the control membrane samples. Significantly down-regulated metabolites are marked in black. Asterisks (*) indicate a statistically significant difference between the control and QSI-modified membranes (*P < 0.05).
The bacterial metabolite content in the biofilm microbiota changed after AC and MA inhibition. The interference of these biochemical pathways (i.e., map00260 and map00520) can inhibit biofilm formation and metabolite (e.g., EPS) biosynthesis. Therefore, introducing inhibitors AC and MA to the membrane reduced bacterial metabolism associated with amino acid and carbohydrate pathways. This resulted in decreased secretion of metabolic substance, such as proteins and polysaccharides, and reduced biofilm formation, effectively mitigating membrane biofouling.
The microbial community composition in seawater varied seasonally, exerting a significant influence on membrane biofouling. This study systematically investigated SWRO membrane biofouling in response to the seasonal fluctuation in microbial communities. The anti-biofouling effectiveness of QSI-functionalized membranes was evaluated along with microbial community analysis. We thus conclude as follows:
Seawater microbial diversity was significantly associated with seasonality, showing higher diversity during warmer weather. Proteobacteria, especially Alpha- and Gammaproteobacteria, dominated the bacterial composition of the membrane biofilm. Interestingly, class Epsilonproteobacteria, despite its less abundance on the membrane, were identified as keystone species within the biofilm, suggesting that some low-abundant bacteria significantly contributed to membrane biofouling.
The relationships between bacteria and biopolymers provided valuable insights for fouling analysis. The relative abundance of polysaccharides within the EPS on the pristine membrane in spring, summer, and fall was higher than that in winter. The higher abundance of EPS-positive-related bacteria, such as Rhodobacteraceae, Vibrio, Acinetobacter, and Cryomorphaceae, emerged as a critical factor contributing to severe membrane biofouling in summer.
AC_m and MA_m membranes exhibited excellent anti-biofouling performances against the seasonality of the microbial community. They mitigated membrane biofouling by suppressing the proliferation of Proteobacteria-affiliated bacteria (e.g., Arcobacter and Rhodobacteraceae), interfering with glycine/serine/threonine and amino/nucleotide sugar metabolism pathways (i.e., map00260 and map00520) and restricting bacterial secretion of polysaccharides and proteins.
Methods
Material and chemicals
Dopamine hydrochloride (98%, stored at 4 °C), glutaraldehyde (GA, 50%, stored at 4 °C), and Tris-HCl (99%) were obtained from Aladdin and used in the membrane modification process. The inhibitors acylase from the porcine kidney (AC) and methyl anthranilate (MA, 99%) were purchased from Sigma Aldrich Inc. and Aladdin Biochemical Technology Co., Ltd., respectively. N-acetyl-l-methionine (Tokyo Chemical Industry, Japan) was prepared by dissolving an appropriate amount of solute in 50 mM phosphate buffer saline (PBS) solution and stored at 4 °C for further use. In the biofouling test, Bacto Marine Broth 2216 (BMB) was purchased from Coolaber Tech Corporation, whereas trace elements solution and vitamin solution were prepared as shown in Supplementary Table 5. Reagents used to characterize the biofilm included sodium hydroxide (NaOH, Sinopharm, 98%), phenol (C6H5OH, Xi-long, 99%), sulfuric acid (H2SO4, Sinopharm, 98%), 96-well plates (Sigma Aldrich), bicinchoninic acid assay kit (BCA Protein Assay Kit, Thermo Scientific, USA), BacTiter-Glo microbial viability assay kit (Promega, USA), Quant-iTTM PicoGreen dsDNA Reagent and Kits (Solarbio), SYTO9 and propidium iodide (LIVE/DEAD Baclight Bacterial Viability Kit, Life Technologies Corporation). Natural seawater (NSW) was collected from Bohai Bay ((38.97N, 117.77E) Tianjin, China), and the water sample was immediately prefiltered through a 0.45-μm microfilter and stored at 4 oC. The commercially available SWRO polyamide membrane, SW30HRLE-8040, was used as the pristine membrane in this study.
Preparation of QSI-modified membranes
Anti-biofouling seawater reverse osmosis (SWRO) membranes were prepared by glutaraldehyde (GA) covalent binding on the polydopamine (PDA)-pretreated SWRO membrane (polyamide membrane)53,54,55 based on Schiff base reaction. Dopamine, as a neurotransmitter, can form a stable polydopamine coating layer via self-polymerization on various solid surfaces. As a crosslinking agent, GA was grafted on the PDA-coated membrane by the reaction between aldehyde groups from GA and hydroxy/amino groups from PDA, which could provide the active site for further covalently binding inhibitors AC and MA. A schematic diagram of QSI immobilization is depicted in Fig. 9, and AC- and MA-modified membranes are designated as AC_m and MA_m, respectively. Specifically, The SWRO membrane was first soaked in 50% ethanol for 2 min and then in deionized water overnight. Subsequently, the membranes were mounted in a custom-made mold with only the active layer exposed for the reaction. Dopamine hydrochloride (100 mL of 2 g L−1) prepared with Tris-HCl buffer (0.05 M, pH 8.5) was poured into the custom-made mold for 1-h coating. Next, 100 mL of 6 wt% GA prepared with phosphate buffer solution (PBS, 50 mM, pH = 7) was added and grafted for 2 h. After removing the unreacted GA with PBS, until no residue reagent remained, 100 mL of 1 mg mL−1 AC (prepared with PBS, 50 mM, pH = 7) and 3 mM MA were added for 12-h QSI immobilization at 4 oC to make AC_m and MA_m membranes, respectively. The membranes prepared were characterized as described in Supplementary Method 1.
Biofouling experiments
The microbial consortia were extracted and enriched from real seawater to evaluate the anti-biofouling performance of the modified membrane56,57. The enrichment method is described in Supplementary Method 2. For the dynamic biofouling experiment, the crossflow SWRO filtration unit with an effective membrane area of 204 cm2 (Supplementary Fig. 9) was washed several times with deionized water, followed by further disinfection by the circulation of 0.5% NaClO for 2 h and recirculation of 75% ethanol for another 1 h. Before the fouling test, the membrane was compacted with deionized water until a constant permeate flux was obtained. After stabilizing the membrane flux, a 100 mL culture of microbial consortia at the exponential growth phase (optical density = 1) was centrifuged and washed in triplicate with phosphate-buffered saline (PBS) solution and subsequently added to the 10-L pretreated NSW (prefiltered by ultrafiltration (PVDF AN50, AMFOR)) before SWRO desalination to achieve the required Silt Density Index (SDI) value for SWRO feedwater) to simulate the open systems of natural desalination fouling conditions. Finally, 0.1% Bacto Marine Broth 2216 (containing 1 ml L−1 of trace element solution and 1 ml L−1 of vitamin solution) (Supplementary Table 1) was added to accelerate the biofouling process57. The following conditions were maintained during the whole experiments: an applied pressure of 6.0 MPa, a temperature of 25 ± 1 oC (using a high-capacity chiller unit, GXDC-0510, China), and a tangential velocity of 0.07–0.135 m s−1. The membrane flux and salt rejection were determined through the PC-interfaced data acquisition system in a crossflow filtration cell. In this study, the winter, spring, summer, and fall seasons are denoted as WI, SP, SU, and FA, respectively. The biofouling experiment was conducted with the pristine membranes (WI_control, SP_control, SU_control and FA_control) and QSI-modified membranes (WI_AC_m, SP_AC_m, SU_AC_m, FA_AC_m, WI_MA_m, SP_MA_m, SU_MA_m and FA_MA_m) under four different season conditions.
Foulant/biofilm extraction and analytical methods
Each fouled RO membrane coupon was cut into several 2 cm × 5 cm segments for foulant extraction. Organic foulants were extracted using NaOH solution (1 M) according to our previous report11. Extraction was done by physical washing with mild sonication (300 W) in 10 mL of extraction solution (contained in 50 mL of carbon-free glass tube) for 30 min and left overnight in the refrigerator. Extracted foulant samples were filtered through a 0.45-μm filters (Millipore) before measurement. The microbial biomass was collected from the fouled membrane and subsequently frozen at −80 °C before being delivered to Majorbio Biopharm Technology Co., Ltd. (Shanghai, China) for high-throughput sequencing42.
The protein and polysaccharide concentrations were determined by the Bradford method (BCA Protein Assay Kit, Thermo Scientific, USA) and phenol-H2SO4 method, respectively. Total organic carbon (TOC) analysis was performed using a TOC-VCSH analyzer (Shimadzu, Boulder, CO, USA). Adenosine triphosphate (ATP) content measurements were carried out using the BacTiter-Glo microbial viability assay kit (Promega, USA) and a 96-well luminometer (Sigma Aldrich) at room temperature for 15 min. The biofilms on the fouled membranes were stained with SYTO9 (ex 480/em 500)/PI (ex 490/em 635) to detect live and dead cell, and were observed using a confocal laser scanning microscope (CLSM) based on the methods of previous studies11. The fluorescence intensity of live cells (500 nm emission (green)) and dead cells (635 nm emission (red)) and biofilm thickness were evaluated by ImageJ. CLSM images were generated using ZEN confocal software, ImageJ software was used to calculate live cells, dead cells, and biofilm thickness total cell fluorescence and was quantified by acquiring z-stacks at an interval of 0.45 µm. A fixed threshold value and connected volume filtration were used for all image stacks.
High-throughput sequencing and data analysis
Next-generation sequencing of the 16 S ribosomal RNA gene was performed for the characterization of microbial community composition and the taxonomic affiliation of the microorganisms extracted from the biofilm growing on the SWRO membrane samples. The detailed procedures are described in the Supplementary Method 3.
Operational taxonomic units (OTUs) with 97% similar cutoff were identified using the UPARSE version 7.1 function within USEARCH58. Poor-quality reads and chimeric sequences were removed using USEARCH default settings. The taxonomy of each OTU representative sequence was classified using the ribosomal database project database. Rarified OTU tables were used to generate alpha and beta diversity metrics59. Bacterial diversity and linear discriminant analysis effect size were both performed using the free online platform of the Majorbio Cloud Platform (www.majorbio.com)60. Bacterial community variation was analyzed with principal coordinate analysis (PCoA) in QIIME2 using unweighted UniFrac distance matrix61 and non-metric multi-dimensional partial least squares discriminant analysis (PLS-DA) as measures of beta diversity. LEfSe analysis was used to compare the significant differences in the bacterial composition between different groups42. A Kruskal–Wallis test was performed to calculate the significance P value in the LEfSe analysis, followed by multiple comparisons adjusted by false discovery rate (fdr). A P value <0.05 indicated a significant difference.
Kyoto Encyclopedia of Genes and Genomes (KEGG) (v2018.01) and Evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) (v4.5) were used for functional annotation and abundance analysis of metabolic pathway. The phylogenetic investigation of communities by reconstruction of unobserved states was used to predict metagenomic functional content based on marker gene data and a database of reference genomes62. Functional genes were selected by searching KOGG to explore the amino acid and carbohydrate mechanisms63.
Phylogenetic molecular ecological networks (pMENs) analysis
pMEN analysis was employed to recognize the principal species and assess their crucial roles inside the bacterial community30,64. Specifically, a module in the Spearman correlation matrices based on the relative abundance of OTU numbers that are highly connected among themselves in pMEN analysis, and the default greedy modularity optimization was selected to separate each network and identify the submodular structure of MENs65. Corresponding random networks were established based on the Maslov–Sneppen method and compared with the pMENs to identify the relative significance of all networks. The statistical analysis of R2 characteristics was applied to describe the networks’ small-world property and modular structure (i.e., the degree of clustering and shortness of paths between nodes). Network visualization was obtained by the Gephi 0.9.2 software (WebAtlas, Paris, France).
Statistical analysis
SPSS statistics25 was used to conduct a one-way analysis of variance (ANOVA) for significance analysis. Asterisks (*) indicate a statistically significant difference between the control and experiment (*P < 0.05). The relationships between bacteria and fouling factors on membrane surfaces were determined using the Spearman correlation heatmap analysis64.
Data availability
Data generated during and/or analyzed during this study are available from the corresponding author on reasonable request. DNA sequences are available at the NCBI Sequence Read Archive, accession number: PRJNA1047337.
Code availability
For access to detailed code implementations, please contact the authors directly.
References
Khan, M. T., Manes, C. L. D., Aubry, C. & Croue, J. P. Source water quality shaping different fouling scenarios in a full-scale desalination plant at the Red Sea. Water Res. 47, 558–568 (2013).
Zhang, Y. et al. Performance fluctuations and evaluation of a piston type integrated high pressure pump-energy recovery device. Npj Clean Water 5, 19 (2022).
Elimelech, M. & Phillip, W. A. The future of seawater desalination: energy, technology, and the environment. Science 333, 712–717 (2011).
Herzberg, M., Kang, S. & Elimelech, M. Role of extracellular polymeric substances (EPS) in biofouling of reverse osmosis membranes. Environ. Sci. Technol. 43, 4393–4398 (2009).
Khan, M. T., Manes, C. L. D., Aubry, C., Gutierrez, L. & Croue, J. P. Kinetic study of seawater reverse osmosis membrane fouling. Environ. Sci. Technol. 47, 10884–10894 (2013).
Al-Juboori, R. A. & Yusaf, T. Biofouling in RO system: mechanisms, monitoring and controlling. Desalination 302, 1–23 (2012).
Kim, T. S. & Park, H. D. Tributyl tetradecyl phosphonium chloride for biofouling control in reverse osmosis processes. Desalination 372, 39–46 (2015).
Kim, T. S. & Park, H. D. Lauroyl arginate ethyl: an effective antibiofouling agent applicable for reverse osmosis processes producing potable water. J. Membr. Sci. 507, 24–33 (2016).
Feng, Y. R. et al. Graphene oxide/methyl anthranilate modified anti-biofouling membrane possesses dual functions of anti-adhesion and quorum quenching. J. Membr. Sci. 668, 121265 (2023).
Turan, N. B., Chormey, D. S., Buyukpinar, C., Engin, G. O. & Bakirdere, S. Quorum sensing: little talks for an effective bacterial coordination. Trend Anal. Chem. 91, 1–11 (2017).
Li, C., Liang, J., Yang, Y., Pu, J. & Hou, L. A. Novel insights into the role of Pseudomonas quinolone signal in the control of reverse osmosis membrane biofouling. J. Membr. Sci. 563, 505–512 (2018).
Li, Y. et al. Anti-biofouling behavior of quorum quenching for removal of pharmaceuticals by forward osmosis membrane based on pseudomonas quinolone signals. J Memb. Sci. 612, 118475 (2020).
Yeon, K. M., Lee, C. H. & Kim, J. Magnetic enzyme carrier for effective biofouling control in the membrane bioreactor based on enzymatic quorum quenching. Environ. Sci. Technol. 43, 7403–7409 (2009).
Ayache, C., Manes, C., Pidou, M., Croue, J. P. & Gernjak, W. Microbial community analysis of fouled reverse osmosis membranes used in water recycling. Water Res. 47, 3291–3299 (2013).
Zodrow, K. R., Bar-Zeev, E., Giannetto, M. J. & Elimelech, M. Biofouling and microbial communities in membrane distillation and reverse osmosis. Environ. Sci. Technol. 48, 13155–13164 (2014).
Flowers, J. J., Cadkin, T. A. & McMahon, K. D. Seasonal bacterial community dynamics in a full-scale enhanced biological phosphorus removal plant. Water Res. 47, 7019–7031 (2013).
Hoek, E. M. V., Weigand, T. M. & Edalat, A. Reverse osmosis membrane biofouling: causes, consequences and countermeasures. Npj Clean Water 5, 45 (2022).
Nguyen, A., Azari, S. & Zou, L. D. Coating zwitterionic amino acid L-DOPA to increase fouling resistance of forward osmosis membrane. Desalination 312, 82–87 (2013).
Shenvi, S. S., Rashid, S. A., Ismail, A. F., Kassim, M. A. & Isloor, A. M. Preparation and characterization of PPEES/chitosan composite nanofiltration membrane. Desalination 315, 135–141 (2013).
Liu, C., Lee, J., Ma, J. & Elimelech, M. Antifouling thin-film composite membranes by cntrolled architecture of Zwitterionic polymer brush layer. Environ. Sci. Technol. 51, 2161–2169 (2017).
Karkhanechi, H., Takagi, R. & Matsuyama, H. Biofouling resistance of reverse osmosis membrane modified with polydopamine. Desalination 336, 87–96 (2014).
Gao, Y. Q. et al. Reverse osmosis membranes with guanidine and amine enriched surface for biofouling and organic fouling control. Desalination 430, 74–85 (2018).
Zhang, S., Zhang, Y. & Chung, T. S. Facile preparation of antifouling hollow fiber membranes for sustainable osmotic power generation. ACS Sustain Chem. Eng. 4, 1154–1160 (2016).
Choudhury, R. R., Gohil, J. M., Mohanty, S. & Nayak, S. K. Antifouling, fouling release and antimicrobial materials for surface modification of reverse osmosis and nanofiltration membranes. J. Mater. Chem. A 6, 313–333 (2018).
Chen, L. et al. Enhancing interface compatibility of UiO-66-NH2 and polyamide by incorporating dopamine into thin film nanocomposite membranes. J Membr. Sci. 654, 120565 (2022).
Levi, A., Bar-Zeev, E., Elifantz, H., Berman, T. & Berman-Frank, I. Characterization of microbial communities in water and biofilms along a large scale SWRO desalination facility: site-specific prerequisite for biofouling treatments. Desalination 378, 44–52 (2016).
Shammi, M., Pan, X. L., Mostofa, K. M. G., Zhang, D. Y. & Liu, C. Q. Seasonal variations and characteristics differences in the fluorescent components of extracellular polymeric substances from mixed biofilms in saline lake. Sci. Bull. 62, 764–766 (2017).
Ying, W., Yang, F., Bick, A., Oron, G. & Herzberg, M. Extracellular polymeric substances (EPS) in a hybrid growth membrane bioreactor (HG-MBR): viscoelastic and adherence characteristics. Environ. Sci. Technol. 44, 8636–8643 (2010).
Vorkapic, D., Pressler, K. & Schild, S. Multifaceted roles of extracellular DNA in bacterial physiology. Curr. Genet. 62, 71–79 (2016).
Wang, H. et al. Membrane autopsy deciphering keystone microorganisms stubborn against online NaOCl cleaning in a full-scale MBR. Water Res. 171, 115390. (2020).
Liu, Y. et al. Effect of magnetic powder on membrane fouling mitigation and microbial community/composition in membrane bioreactors (MBRs) for municipal wastewater treatment. Bioresour. Technol. 249, 377–385 (2018).
Guo, X. C. et al. Correlation between microbial community structure and biofouling as determined by analysis of microbial community dynamics. Bioresour. Technol. 197, 99–105 (2015).
Zhu, L. F. et al. Environmental temperatures affect the gastrointestinal microbes of the Chinese giant salamander. Front. Microbiol. 12, 543767 (2021).
Rehman, Z. U., Ali, M., Iftikhar, H. & Leiknes, T. Genome-resolved metagenomic analysis reveals roles of microbial community members in full-scale seawater reverse osmosis plant. Water Res. 149, 263–271 (2019).
Chen, C. et al. A comprehensive evaluation of the temporal and spatial fouling characteristics of RO membranes in a full-scale seawater desalination plant. Water Res. 249, 120914 (2023).
Jiang, L. J., Chen, L. & Zhu, L. Fouling process of membrane distillation for seawater desalination: an especial focus on the thermal-effect and concentrating-effect during biofouling. Desalination 485, 114457 (2020).
Nagaraj, V., Skillman, L., Ho, G., Li, D. & Gofton, A. Characterisation and comparison of bacterial communities on reverse osmosis membranes of a full-scale desalination plant by bacterial 16S rRNA gene metabarcoding. Npj Biofilms Microbiol. 3, 13 (2017).
Kim, L. H. & Chong, T. H. Physiological responses of salinity-stressed vibrio sp and the effect on the biofilm formation on a nanofiltration membrane. Environ. Sci. Technol. 51, 1249–1258 (2017).
Fu, H. M., Wang, J. F., Ren, H. Q. & Ding, L. L. Acceleration of start-up of moving bed biofilm reactor at low temperature by adding specialized quorum sensing bacteria. Bioresour. Technol. 358, 127249 (2022).
Deng, L. J. et al. Application of a specific membrane fouling control enhancer in membrane bioreactor for real municipal wastewater treatment: sludge characteristics and microbial community. Bioresour. Technol. 312, 123612 (2020).
Flemming, H. C. et al. The biofilm matrix: multitasking in a shared space. Nat. Rev. Microbiol. 21, 70–86 (2022).
Liu, C., Zhu, L. & Chen, L. Effect of salt and metal accumulation on performance of membrane distillation system and microbial community succession in membrane biofilms. Water Res. 177, 115805 (2020).
Perez-Rodriguez, I., Bolognini, M., Ricci, J., Bini, E. & Vetriani, C. From deep-sea volcanoes to human pathogens: a conserved quorum-sensing signal in Epsilonproteobacteria. ISME J. 9, 1222–1234 (2015).
Doberva, M. et al. Large diversity and original structures of acyl-homoserine lactones in strain MOLA 401, a marine Rhodobacteraceae bacterium. Front. Microbiol. 8, 1–10 (2017).
Zhang, Z. M. et al. Understanding of aerobic sludge granulation enhanced by sludge retention time in the aspect of quorum sensing. Bioresour. Technol. 272, 226–234 (2019).
Xiong, F. Z., Zhao, X. X., Wen, D. H. & Li, Q. L. Effects of N-acyl-homoserine lactones-based quorum sensing on biofilm formation, sludge characteristics, and bacterial community during the start-up of bioaugmented reactors. Sci. Total Environ. 735, 139449 (2020).
Mayer, C., Romero, M., Muras, A. & Otero, A. Aii20J, a wide-spectrum thermostable-acylhomoserine lactonase from the marine bacterium sp 20J, can quench AHL-mediated acid resistance. Appl. Microbiol. Biotechnol. 99, 9523–9539 (2015).
Vesuna, A. P. & Nerurkar, A. S. Biocontrol impact of AHL degrading actinobacteria on quorum sensing regulated virulence of phytopathogen Pectobacterium carotovorumsubsp.carotovorumBR1. Plant Soil 453, 371–388 (2020).
Xu, B., Cho, Q. A. C., Ng, T. C. A., Huang, S. & Ng, H. Y. Enriched autoinducer-2 (AI-2)-based quorum quenching consortium in a ceramic anaerobic membrane bioreactor (AnMBR) for biofouling retardation. Water Res. 214, 118203 (2022).
Feng, Y., Zhao, Y. P., Guo, Y. Z. & Liu, S. T. Microbial transcript and metabolome analysis uncover discrepant metabolic pathways in autotrophic and mixotrophic anammox consortia. Water Res. 128, 402–411 (2018).
Papenfort, K. & Bassler, B. L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588 (2016).
Xing, A. Q. & Last, R. L. A regulatory hierarchy of the Arabidopsis branched-chain amino acid metabolic network. Plant Cell 29, 1480–1499 (2017).
Zhang, H. R., Luo, J. Q., Li, S. S., Wei, Y. P. & Wan, Y. H. Biocatalytic membrane based on polydopamine coating: a platform for studying immobilization mechanisms. Langmuir 34, 2585–2594 (2018).
Li, X., Xu, Y. L., Goh, K., Chong, T. H. & Wang, R. Layer-by-layer assembly based low pressure biocatalytic nanofiltration membranes for micropollutants removal. J. Membr. Sci. 615, 118514 (2020).
Kim, T. S., Park, S. H., Park, D., Lee, J. H. & Kang, S. Surface immobilization of chlorhexidine on a reverse osmosis membrane for in-situ biofouling control. J. Membr. Sci. 576, 17–25 (2019).
Ansari, A., Pena-Bahamonde, J., Fanourakis, S. K., Hu, Y. D. & Rodrigues, D. F. Microbially-induced mineral scaling in desalination conditions: mechanisms and effects of commercial antiscalants. Water Res. 179, 115863 (2020).
Ansari, A. et al. Polyacrylic acid-brushes tethered to graphene oxide membrane coating for scaling and biofouling mitigation on reverse osmosis membranes. J. Membr. Sci. 630, 119308 (2021).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996 (2013).
Belila, A. et al. Bacterial community structure and variation in a full-scale seawater desalination plant for drinking water production. Water Res. 94, 62–72 (2016).
Ma, L., Yang, Y., Liu, W. H. & Bu, D. P. Sodium butyrate supplementation impacts the gastrointestinal bacteria of dairy calves before weaning. Appl. Microbiol. Biotechnol. 107, 3291–3304 (2023).
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J. & Knight, R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172 (2011).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814 (2013).
Feng, J. Y. et al. Enhanced permeation performance of biofiltration-facilitated gravity-driven membrane (GDM) systems by in-situ application of UV and VUV: comprehensive insights from thermodynamic and multi-omics perspectives. Water Res. 242, 120254 (2023).
Zhu, X. Z. et al. Deciphering mono/multivalent draw solute-induced microbial ecology and membrane fouling in anaerobic osmotic membrane bioreactor. Water Res. 209, 117869 (2022).
Wang, Z. Y. et al. Network analysis reveals microbe-mediated impacts of aeration on deep sediment layer microbial communities. Front. Microbiol. 13, 931585 (2022).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 52070018 and Grant NO. 51920105012) and the National Key Research and Development Program of China (Grant No. 2021YFC3201403).
Author information
Authors and Affiliations
Contributions
Chao Chen: Conceptualization, methodology, writing—original draft preparation, writing—review & editing. Yu Yang: Supervision, project administration, funding acquisition, writing—review & editing. Kwang-Ho Choo: Conceptualization, validation, writing—review & editing. How Yong Ng: Validation, writing—review & editing. Satoshi Takizawa: Validation, writing—review & editing. Li-an Hou: Supervision, funding acquisition, writing—review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, C., Yang, Y., Choo, KH. et al. Cracking the code of seasonal seawater biofouling: enhanced biofouling control with quorum sensing inhibitor-functionalized membranes. npj Clean Water 7, 12 (2024). https://doi.org/10.1038/s41545-024-00305-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41545-024-00305-w