Abstract
Antimicrobial resistome in wastewater treatment plants was investigated via shotgun metagenomic analysis over a variety of geographical locations, seasons, and biological treatment configurations. The results revealed that the transition of the antimicrobial resistome occurred at two locations during wastewater treatment, which resulted in a distinctive antimicrobial resistome in influent wastewater, activated sludge, and treated effluent. The antimicrobial resistome in influent wastewater was characterized by a high abundance of antibiotic resistance genes (ARGs) on clinically important drugs, whereas sludge retained a higher abundance of multidrug ARGs associated with efflux pump. Seasonality was the primary factor affecting antimicrobial resistome in influent wastewater, which partially succeeded to the subsequent resistome of activated sludge and treated effluent. Importantly, some ARGs on clinically important drugs in influent wastewater passed through the biological treatment to be discharged in the treated effluent, except in the membrane bioreactor process.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR) is a major threat to human health. A recent study reported that AMR is now the leading cause of death in the world, and it was associated with 1.3 million deaths in 20191. According to O’ Neil2, the health burden caused by AMR will continue to increase, resulting in 10 million annual deaths in 2050. To prevent the spread of AMR, the “One Health” approach is urgently needed, which is a collaborative effort by multiple disciplines to attain optimal health for people, animals, and the environment3. Among the environmental pathways of AMR spread, wastewater has been regarded as the primary source of the emergence of AMR in the water environment. Particularly, the discharge of untreated wastewater has been reported to result in a significant increase in AMR prevalence in the water environment in various countries4,5,6,7,8,9. Accordingly, wastewater treatment plants (WWTPs) have emerged as an important barrier to prevent the spread of AMR in wastewater into the water environment. However, WWTPs can also act as AMR reservoirs, retaining it in the activated sludge and potentially discharging it into treated effluent10,11,12,13. Particularly, studies have revealed that WWTP effluent still contains a certain abundance of antimicrobial-resistant bacteria (ARB) and antimicrobial resistance genes (ARGs)14,15,16,17. Hence, treated effluent from WWTPs can be another major source of AMR in water bodies and drinking water resources receiving the treated effluent18,19,20,21.
Although both wastewater and treated effluent are the major anthropogenic sources of AMR in water environments, wastewater, and treated effluent may exhibit different AMR traits. The major source of AMR in wastewater is the gut microbiome, wherein the AMR is enriched via exposure to antimicrobial doses from people in the sewershed community. In contrast, AMR in treated effluent mainly originates from activated sludge, which is exposed to a variety of stresses that induce the coselection and enhancement of AMR (e.g., heavy metals, reactive oxygen species (ROS), disinfectants, and sub-lethal concentrations of antimicrobials)22,23,24. Hence, the discharge of AMR via wastewater or treated effluent poses a qualitatively different impact on water environment. Several studies have demonstrated the difference in the antimicrobial resistome of influent wastewater and treated effluent using the shotgun metagenomic approach25,26,27. These studies have provided fundamental insights through case studies by targeting a single WWTP; however, the factors that generally govern the fate of antimicrobial resistome among various WWTPs with different biological process configuration, and geographic location are yet to be comprehensively understood. Moreover, these studies analyzed the shotgun metagenomic sequence data based on raw reads of the observed ARGs, which are highly biased compared to the original proportion of each ARG, because a longer gene appears to have greater numbers of gene fragments in shotgun metagenomic sequencing. For example, mexB, which exhibit multiple resistance, and tetQ, which exhibit tetracycline resistance, are 3141- and 1974-bp long, respectively, whereas mphB, which exhibits macrolide resistance, and qnrS, which exhibit fluoroquinolone resistance, are 477 and 657 bp long, respectively. This could result in five–tenfold overestimations in the proportion of long ARGs in a raw reads-based quantification. Therefore, the appropriate normalization of raw reads is essential to acquiring accurate snapshots of the antimicrobial resistome and understanding the mechanism and the factors involved in the transition of antimicrobial resistome from influent wastewater to treated effluent in WWTPs.
This study aimed to reveal the key factors governing the transition of antimicrobial resistome during wastewater treatment processes. Accordingly, the antimicrobial resistome in influent wastewater, activated sludge, and treated effluent was investigated using systematic sampling and shotgun metagenomic analysis over a variety of geographical locations, seasons, and biological treatment configurations. In this study, all the ARG data were curated after normalization as reads per kilobase (RPK) to obtain an accurate snapshot of the antimicrobial resistome, which reflects the original proportion of the gene copy number in a sample. In addition, the common characteristics and differences in the antimicrobial resistome (i.e., ARG abundance and composition) in influent wastewater, activated sludge, and treated effluent, were identified by introducing an appropriate normalization. Moreover, regarding the differences among the resistome and their association with the microbial community, this study revealed the impact of key factors influencing the antimicrobial resistome and its transition during wastewater treatment processes.
Results and discussion
ARG abundance in the wastewater treatment process
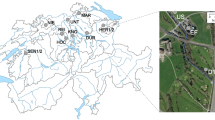
Influent wastewater, activated sludge, and treated effluent were collected from five municipal WWTPs with different biological treatment process configuration (Fig. 1). The total ARG abundance per 16 S bacterial population in influent wastewater substantially decreased in activated sludge, regardless of the process configuration and geographical location of the WWTPs. The total ARG abundance in influent wastewater from all the WWTPs was 32–50% (Fig. 2a). No remarkable difference in the total ARG abundance in influent wastewater among the WWTPs located in different municipalities was observed. The total ARG abundance in the activated sludge collected from all the WWTPs decreased substantially to 5–19%. This decrease from influent wastewater to activated sludge was observed in all samples regardless of the season or process configuration. A slight increase or no remarkable change in the total ARG abundance was observed from sludge to the final effluent collected from all WWTPs, although the reduction in the bacterial population was substantial. The ARG abundances observed in this study are consistent with the typical range of total ARG abundance reported in the literature based on the metagenomic approach, which was reported as 20–40%28,29. Similarly, the decrease in the ARG abundance from influent wastewater to activated sludge was reported in previous studies. The metagenome-based total ARG abundance in activated sludge was reported to decrease by 12–13% from influent wastewater to 8–9%28. Further, studies based on high-throughput qPCR reported a 10–20% decrease in the abundance of major ARGs from influent to sludge15,17. However, several studies on culture-based AMR assays have reported no remarkable change or only a slight increase in the AMR abundance in sludge and final effluent compared to those in influent wastewater14,30,31,32,33. This is probably because the culture-based assays target only target specific bacteria groups of fecal origin (e.g., coliform, E. coli, and Salmonella), whereas metagenomic approaches cover the entire bacterial community. Observation of the total ARG abundance at each treatment stage revealed that the ARG abundance decreased during biological treatment (Fig. 2b). However, no change in the total ARG abundance from influent wastewater was observed after primary sedimentation until the equalization tank. This indicates that primary sedimentation and retention in the equalization tank had no apparent effect on the ARG abundance in influent wastewater. The total ARG abundance decreased to 15–21% during the CAS process and 10–12% during the MBR process when the primary-treated wastewater entered the biological treatment stage.
a Influent wastewater (WW), activated sludge (SL), and treated effluent (EFF) from various WWTPs with different process configurations. b Each treatment stage of conventional activated sludge (CAS) and membrane bioreactor (MBR) processes at WWTP B. WW influent wastewater, PTE primary sedimentation effluent, EQ equalization tank, Ano (MBR) anoxic tank sludge from the MBR process, Ox (MBR) aeration tank sludge from the MBR process, Mem (MBR) membrane filtration tank sludge from the MBR process, Ox (CAS) aeration tank sludge from the CAS process, STE (CAS) final sedimentation effluent from the CAS process, STE (MBR) membrane permeate from the MBR process, EFF final chlorinated effluent from WWTP B.
The total ARG abundances in the treated effluent were similar to those in the activated sludge and apparently lower than those in influent wastewater (Fig. 2). Although the 16 S bacterial population in the treated effluent reduced by 1.0–1.7 log compared to that in sludge, no remarkable change in the total ARG abundance was observed. These trends in the ARG abundance were consistent with those reported in previous studies17,28. The change in the total ARG abundance before and after chlorination was not remarkable (Fig. 2b). Previous studies have reported that the log reduction in ARGs in secondary treated effluent is limited under the typical chlorination intensity of 15 mg × min L−1 (see refs. 34,35,36,37), although the population of ARB is substantially reduced. Similarly, in the present study, chlorination reduced the total bacteria population; however, the impacts of chlorination on the reduction in ARG abundance were limited.
In accordance with total ARG abundance, the ARG composition also exhibited a remarkable change from influent wastewater to activated sludge. Only 45–50% of ARGs in influent wastewater remained in activated sludge, and 84–91% of the remaining ARGs in the sludge were carried over to the treated effluent (Fig. 3). The proportion of multiple antimicrobial resistance (MAR) genes in the sludge increased, regardless of the process configuration and location of WWTPs (Fig. 4). Particularly, the proportion of ARGs with broad resistance to more than six drug classes increased notably in the activated sludge compared to the influent wastewater. This remarkable change in ARG abundance and composition from wastewater to sludge indicates that activated sludge maintained its unique antimicrobial resistome. During biological treatment, part of the activated sludge was returned to the beginning of the biological treatment after thickening in the final sedimentation tank. The returned thickened sludge (namely return sludge) mixed with the primary-treated wastewater to constitute the activated sludge. Under the typical operating conditions of WWTPs, the flow rate of the return sludge is configurated as 1/2–3/4 of the flow rate of the primary-treated wastewater38. Meanwhile, the suspended solids (SS) concentration of the return sludge is typically thickened to 6000–12,000 mg L−1, which is 3–5 times higher than that of activated sludge in the aeration tank. Hence, most of the biosolids in the activated sludge originated from the return sludge. The estimated biosolids ratio of primary-treated wastewater to return sludge in the aeration tank is 1:200–1000. The flow rate of the return sludge in WWTP B was 25–40% of that of the primary-treated effluent, whereas the SS in the return sludge was 3.4–4.4 times higher than that in activated sludge. Thus, the antimicrobial resistome in activated sludge was mainly comprised of those in the return sludge rather than those in influent wastewater. Consequently, the impacts of influent wastewater on the antimicrobial resistome in the activated sludge was relatively small.
Seasonal differences were also observed for both ARG abundance and composition. The increase in the ARGs with broad resistance (<6) was more notable in summer (+ 0.9–6.5%) than in winter (+ 1.5–11%). In contrast, the proportion of ARGs resistant to two drug classes was remarkably higher in winter. These results indicated the seasonality of ARG compositions in influent wastewater and activated sludge. In addition, the ARG proportion resistant to each drug class differed between influent wastewater and activated sludge (Fig. 5). Particularly, compared to the sludge samples, the influent wastewater samples in all WWTPs were mostly composed of ARGs resistant to tetracycline, fluoroquinolone, and macrolide. Moreover, seasonality was observed in the proportion of ARGs resistant to some drug classes. For example, the proportion of ARGs resistant to macrolide and streptogramin was larger in winter, whereas those resistant to aminoglycoside exhibited a large proportion regardless of the season. The comparison of the total ARG abundance and AMR compositions revealed that the following characteristics in the antimicrobial resistome in the WWTPs: (i) the total ARG abundance in influent wastewater was ~30–50% regardless of season and geographical location of WWTPs, (ii) the total ARG abundance substantially reduced in the sludge regardless of the process configuration, (iii) ARG composition and proportion of MAR changed from influent wastewater to sludge and with season, (iv) chlorination substantially reduced bacterial population but had no marked effect on the abundance of AMR.
a ARGs to a single drug class in influent wastewater in winter, b ARGs to two drug classes in influent wastewater in winter, c ARGs to a single drug class in influent wastewater in summer, d ARGs to two drug classes in influent wastewater in summer, e ARGs to a single drug class in activated sludge in winter, f ARGs to two drug classes in activated sludge in winter, g ARGs to single drug class in activated sludge in summer, h ARGs to two drug classes in activated sludge in summer.
Transition of ARG composition during wastewater treatment
The ARG composition at five WWTPs was compared via principal component analysis (PCA) (Fig. 6a). The primary factor distinguishing ARG compositions was the treatment stage (i.e., influent wastewater, activated sludge, and treated effluent), whereas seasonality was the secondary factor. In contrast, geographic location and process configuration of WWTPs had no apparent effect on the ARG compositions in influent wastewater and activated sludge. The influent wastewater was mostly characterized by negative PC1 scores except for WWTP E in summer, whose influent wastewater may contain some sludge due to sampling restrictions (as described in “Methods”). The negative PC1 values indicated the influence of AMR on critically important antimicrobials, including quinolones (qnrS, qnrVC), macrolides (ermG, mel), aminoglycosides (APH(3’), cmlA, aad(6), AAC(6’)), colistins (ICR), cephalosporins, and carbapenem (ESBL genes of MOX, OXA, CfxA, VEB, FOX, GES, and CMY) (Fig. 6a and Supplementary Table 4a). In contrast, the activated sludge samples were characterized by MAR genes, which exhibited highly positive PC1 scores (Fig. 6a). Among the top 30 ARGs with highly positive PC1 loadings, 19 ARGs were efflux pumps associated with broad resistance to five or more antibiotic classes (mexD, medQ, mexW, mexI, mexK, mexN, MuxC, MucB, and mexF) (Supplementary Table 4b). Typically, the efflux pump functions as a resistance to not only antimicrobials but also a broad range of cell stresses. During biological treatment, sludge bacteria are exposed to various kinds of cell stresses (e.g., antimicrobials, heavy metals, toxic trace chemicals, and oxidative stress by aeration). In an aeration tank, these cell stresses may result in the enhancement or coselection of antimicrobial resistance23,24,39,40,41,42. Interestingly, the treated effluents were plotted at an intermediate range between influent wastewater and sludge groups (Fig. 6a). Treated effluent is expected to exhibit a similar ARG composition as sludge if each ARG in sludge is equally reduced in the final sedimentation. However, this PCA result demonstrated that the ARG compositions in treated effluent were not solely affected by activated sludge but also by influent wastewater. This is consistent with the findings that the ARGs originating from influent wastewater partly remained in the final effluent of the WWTP (Fig. 3). Some ARGs were not detected in activated sludge, although they were found both in influent wastewater and treated effluent, as shown in Fig. 3. Bacterial DNA originating from wastewater were present in activated sludge at lower abundances than that originating from the return sludge. Hence, some ARGs of wastewater origin were present but at a lower abundance than the detection threshold, which was ~1 per 30 million reads.
a ARG composition in influent wastewater, activated sludge, and treated effluent at various WWTPs with different process configurations. b ARG composition in each treatment stage of CAS and MBR processes at WWTP B. c 16S-based microbial community in influent wastewater, activated sludge, and treated effluent at various WWTPs with different process configurations. d 16S-based microbial community in each treatment stage of CAS and MBR processes at WWTP B.
The secondary factor affecting ARG composition was season, which was represented by PC2 (Fig. 6a). Samples collected in winter exhibited positive PC2 scores, which were characterized by ARGs of antibiotic inactivation associated with resistance to β-lactams, such as cephalosporins; carbapenem; and ESBL-producing genes of OXA, MOX, GES, SHV, CMY, and ADC (Supplementary Table 5a). In contrast, samples collected in summer mostly exhibited negative PC2 scores, which were mainly characterized by ARGs of efflux pumps associated with MAR. The seasonal difference in ARG composition was larger in influent wastewater than in activated sludge. This may be attributed to the seasonal use of clinical antimicrobials in Japan (as discussed below). Treated effluents in winter and summer were also distinguished by PC2. The PC2 scores of the treated effluents in winter and summer were intermediate ranging between those of influent wastewater and activated sludge in winter and summer, respectively. This supports the hypothesis that the treated effluent was not only affected by activated sludge but also by influent wastewater.
The shift in ARG composition occurred at two locations during the course of the treatment process according to the PCA scores in each treatment stage of WWTP B (Fig. 6b). No notable change occurred in the ARG compositions in the influent wastewater (WW) even after primary sedimentation until equalization tank. However, the PC1 scores turned positive when the primary-treated wastewater was subjected to biological treatment. All the sludge in the biological treatment of both CAS and MBR exhibited similar ARG compositions that were associated with highly positive PC1 scores. These results indicated that ARG composition remarkably changed at the beginning of biological treatment. This shift in the ARG composition is consistent with the shift in the total ARG abundance and MAR proportion during biological treatment (Figs. 2 and 4). During the CAS process, a second shift in the ARG composition was observed in the secondary treated effluent after final sedimentation (STE(CAS)). CAS effluent was plotted at an intermediate range between those of the wastewater and sludge groups, suggesting that the ARG composition of the CAS effluents was not only affected by sludge but also by wastewater. This indicated that ARGs of wastewater origin were partly retained in the CAS effluent. In MBR, however, no remarkable change from sludge to MBR effluent was observed. MBR effluents exhibited positive PC1 scores, indicating that their ARG compositions remained similar to those of sludge. This demonstrated that the second shift in the ARG compositions occurred in CAS at final sedimentation but not in MBR, in which the sludge was eliminated via membrane filtration. These differences between CAS and MBR effluents demonstrated that the ARG composition in the treated effluent was affected by the process configuration, particularly at the sludge separation stage. Moreover, the ARGs of wastewater origin were likely substantially reduced through membrane filtration in the MBR, whereas their reduction was limited in final sedimentation in CAS. The second shift in the ARG composition in CAS likely occurred via the overflow of ARGs of wastewater origin at the final sedimentation.
Association of microbial community with ARG composition
Some phylogenetic classes were found to be associated with specific ARG groups during wastewater treatment. The relative abundance of Bacteroidia and Flavobacteriia classes in Bacteridetes, Bacilli, Clostridia, and Erysipelotrichi classes in Firmicutes, Fusobacteriia, Eplilonproteobacteria, Gammaproteobacteria, and Synergistia classes (hereinafter referred to as microbial group A) had relatively high correlations with some ARGs to macrolide (ErmA, ErmB, ErmF, ErmG, mefC, mefE, and mel), quinolone (QnrD and QnrS), tetracycline (tet3, tetM, tetO, tetQ, tetS, and tetW) and vancomycin (vanB, vanW, vanX, and vanY), whereas microbial group A had relatively low correlations with most ARGs to sulfonamide (sul1, sul2, and sul4) and with multidrug ARGs of the efflux pump (most of the mex and Mux genes) (Fig. 7). Microbial group A mostly consisted of phylogenetic classes for commensal bacteria and anaerobic bacteria which are often abundant in the gut microbiome. These bacteria were abundant in influent wastewater and possibly harbored ARGs to macrolide, quinolone, tetracycline, and vancomycin in influent wastewater. Interestingly, some phylogenetic classes (hereinafter referred to as microbial group C) had the opposite trend of correlations to microbial group A. Microbial group C mostly consisted of phylogenetic classes, which are often associated with water and soil environments, namely Alphaproteobacteria, Deltaproteobacteria, Cytophagia, and Nitrospira and classes in Acidobacteria, Actinobacteria, Chloroflexi, Planctomycetes, and Verrumicrobia phyla, etc., which were abundant in the sludge samples (Supplementary Table 6b). Microbial group C had relatively high correlations with ARGs to sulfonamide (sul1, sul2, and sul4) and to multiple antimicrobials by efflux (most of the mex and Mux genes) (Fig. 7). Hence, these microbes in activated sludge possibly harbored and reserved the efflux pump ARGs with broad resistance in activated sludge.
As observed in the ARG composition, the microbial community was differed among the influent wastewater, activated sludge, and treated effluent samples (Fig. 6c). Unlike the ARG composition, however, the seasonal change did not exhibit a remarkable effect on the microbial community. In contrast, process configuration exhibited a greater impact on the microbial community in the sludge samples. The influent wastewater samples exhibited negative PC1 scores, whereas the activated sludge samples exhibited highly positive PC1 scores. The microbial community of influent wastewater was characterized by microbes with highly negative PC1 loadings, which were mainly composed of anaerobic enteric bacteria, such as Bacteroidetes, Firmicutes, and Gammaproteobacteria, including Enterobacteriaceae and Aeromonadaceae (Supplementary Table 6a). In contrast, the sludge samples were characterized by microbes with highly positive PC1 loadings, which largely consisted of aerobic bacteria, including Deltaproteobacteria and Planctomycetia (Supplementary Table 6b). Similarly, the PC1 scores of the treated effluent were plotted at an intermediate range: between those of influent wastewater and activated sludge, as observed in the ARG composition. This suggested that the microbial community in the treated effluent was not only affected by activated sludge but also by influent wastewater. Hence, some microbes in influent wastewater were not reduced but retained in the final effluent of WWTPs. If these microbes harbored ARGs and remained in the treated effluent, the ARG composition in the treated effluent would be partly dependent on the influent wastewater characteristics. In the present study, most of the microbes characterizing the influent wastewater belonged to microbial group A, which had high correlations with ARGs to macrolide, quinolone, tetracycline, and vancomycin and was found abundantly in influent wastewater (Fig. 7). These bacteria were potential carriers of ARGs to clinically important drugs remaining in the treated effluent.
Transition of the microbial community also occurred at two locations during the treatment process, as observed in the ARG compositions (Fig. 6d). The first shift occurred when primary-treated wastewater was subjected to biological treatment, during which the PC1 scores turned from negative to positive. The sludge group was divided into CAS and MBR groups, which were characterized by negative and positive PC2 scores, respectively. The second shift in the microbial community occurred at the sludge separation stage after biological treatment. Treated effluents exhibited intermediate PC1 scores of those of the wastewater and sludge groups. Therefore, the shift in the microbial community corresponded with that of the ARG composition. Importantly, the second shift in the microbial community after the sludge separation stage was observed in both CAS and MBR. This indicates that process configuration, particularly sludge separation, had no remarkable effect on the microbial community of the treated effluent but had a notable effect on the ARG composition.
In summary, the microbial community and antimicrobial resistome had the following relations during wastewater treatment: (i) anaerobic enteric bacteria abundant in influent wastewater were associated with ARGs to macrolide, fluoroquinolone and tetracycline; (ii) the microbial community drastically changed at biological treatment, during which the microbes were associated with multidrug ARG became more abundant; (iii) the microbial community in CAS effluent and MBR effluent did not differ remarkably, whereas antimicrobial resistomes were distinctively different.
Determinative factors of antimicrobial resistome in WWTPs
The transition in the antimicrobial resistome in the wastewater treatment process exhibited a common tendency regardless of the geographical locations of the WWTPs. In summary, the transition in the ARG composition occurred at two locations during the course of the wastewater treatment process. Consequently, distinctive ARG compositions were developed in the influent wastewater, activated sludge, and treated effluent. In the present study, seasonality was the primary factor characterizing the “wastewater resistome” (i.e., antimicrobial resistome in influent wastewater and primary-treated wastewater). The wastewater resistome is likely to reflect the clinical use of antimicrobials in the sewershed community. Thus, the wastewater resistome was affected by seasonality in antimicrobial use. The wastewater resistome was mainly characterized by ARGs resistant to aminoglycoside, cephalosporins, macrolides, quinolones, and tetracyclines (Fig. 5 and Supplementary Table 4a), which are commonly used antimicrobials in clinics in Japan43 (Fig. 8). In Japan, the prescription of 3rd-generation cephalosporins, macrolides and other antimicrobials has increases seasonally in winter (Dec–Feb)43,44,45. Accordingly, the abundance of ARGs to cephalosporins and macrolides increased in the wastewater resistome (Fig. 5 and Supplementary Table 5a). This seasonality in the wastewater resistome was likely partially succeeded to the subsequent resistome of activated sludge and treated effluent in accordance with its dependence on the wastewater resistome.
The “sludge resistome” was less diverse and had no apparent determinative factors except the seasonality succeeded from influent wastewater. The sludge resistome was not markedly affected by the process configuration, operating parameters nor geographical location of the WWTPs, although higher BOD in wastewater and short sludge retention time (SRT) possibly affected the difference of sludge resistome among the WWTPs only in summer (Supplementary Fig. 1 and Supplementary Table 1). Hence, the unique resistome was developed in activated sludge while sludge was retained in the biological treatment system via the circulation of return sludge. In the biological treatment, microbes which harbor resistance genes to broad virulence could be gradually enriched by coselection through exposure to various virulent substances while being circulated and retained in the biological treatment. Consequently, all the observed resistomes in activated sludge were characterized by a higher abundance of ARGs associated with a broad AMR under the efflux pump (Supplementary Table 4b). A high abundance of multidrug ARGs in sludge was also reported in past studies26,27. Ng et al.26 reported that sludge in MBR exhibits a higher abundance of multidrug ARGs of the mex gene family, which exhibit broad resistance to most drug classes. The increased abundance of the mex gene family in the “sludge resistome” was also observed in the present study. Hence, activated sludge is an important reservoir and potential source of ARGs with multiple resistances, which may be discharged from WWTP effluent into water environment.
The primary factor affecting the “effluent resistome” was the process configuration of the sludge separation stage. The second shift in the ARG composition from “sludge resistome” to “effluent resistome” occurred during the final sedimentation. However, this second shift was not observed in MBR, suggesting that the configuration of sludge separation is the key factor characterizing the “effluent resistome”. Understanding the resistome in treated effluent is essential because WWTPs are an important barrier for preventing the spread of AMR into the environment. Past studies reported that treated effluent had a different antimicrobial resistome from that of influent wastewater46,47. However, the present study revealed that the resistome of treated effluent is not fully independent of that of influent wastewater. On the contrary, ~90% of ARGs in treated effluent were common to influent wastewater (Fig. 3). These facts demonstrated that a certain portion of ARGs in influent wastewater bypassed biological treatment and final sedimentation to be retained in the treated effluent. Raza et al.48 reported that some ARGs commonly present in influent wastewater and treated effluent were associated with opportunistic pathogenic bacteria. In this study, the phylogenetic classes, including opportunistic pathogenic bacteria, were associated with ARGs of wastewater origin. These findings also support that ARGs of wastewater origin possibly passed through the wastewater treatment processes and remain in the effluent. In contrast, the MBR effluent had a similar resistome to sludge, which is markedly different from the wastewater resistome. This suggests that sludge separation is the key step to excluding ARGs of wastewater origin from treated effluent. Membrane filtration in MBR achieves a good reduction of microbes and extracellular ARGs in the supernatant, whereas the final sedimentation tank is mechanistically unable to eliminate microbes and extracellular ARGs which are present in the supernatant. Therefore, the independence of the resistome in the MBR effluent of the wastewater resistome implies that ARGs of wastewater origin were abundant in the supernatant of activated sludge. In fact, extracellular ARGs are reportedly abundant in the treated effluent of CAS49. Eliminating ARGs in the supernatant of activated sludge is important to reduce ARGs of wastewater origin in the treated effluent. The results of this study suggested that application of MBR could be effective for reducing ARGs of wastewater origin discharged via WWTP effluent. Actually, MBR reportedly exhibited a good reduction in extracellular genes even with an MF membrane due to cake layer filtration50. In this study, the effluent resistome from conventional final sedimentation was not only composed of ARGs from sludge but also ARGs from influent wastewater. Hence, treated effluent is a potential source of a large variety of ARGs, including both multidrug ARGs with broad AMR from activated sludge as well as ARGs to clinically important drugs from influent wastewater. Therefore, reducing ARG in WWTP effluent is essential to reducing the variety of ARGs discharged into the water environment.
In this study, remarkable difference and seasonal change of antimicrobial resistome in influent wastewater, activated sludge and treated effluent were revealed. However, the statistical comparison could not be performed because seasonal composite samples were used in the present study. Antimicrobial resistome in influent wastewater, activated sludge and treated effluent have hourly change and daily fluctuation caused by various environmental factors including sewershed and operating conditions. Further studies are expected on the fluctuations of antimicrobial resistome during wastewater treatment and factors causing the fluctuations for more mechanistic understanding on fate of antimicrobial resistance in WWTPs. Despite these limitations, WWTPs were likely to reserve a unique resistome, which is distinctive from resistome in wastewater, independent of geographical locations. This finding demonstrated that a WWTP is potentially a unique and important reservoir of AMR in the environmental domain in any country. Uniformity (or difference) of antimicrobial resistome in WWTPs among different countries is expected to be examined further for better understanding of the role of WWTPs as the source and reservoir of AMR. More importantly, this study demonstrated that conventional wastewater treatment processes are not optimized to reduce ARGs of wastewater origin, which abundantly compose ARGs to clinically important antimicrobials. Development of novel process design and optimization of operating conditions are expected to enhance the role of a WWTP as a barrier to prevent the spread of AMR from wastewater into aquatic environment.
Methods
Sampling at wastewater treatment plants
Influent wastewater, activated sludge, and treated effluent samples were collected at five municipal WWTPs located in different municipalities in Japan. The target WWTPs were selected to ensure variation in the biological treatment process configuration: conventional activated sludge (CAS) process at WWTP A, CAS and MBR processes treating identical wastewater in parallel at WWTP B, enhanced biological phosphorus removal (EBPR) process at WWTP C, anaerobic–anoxic–oxic (A2O) process at WWTP D, and oxidation–ditch (OD) process at WWTP E (Fig. 1). The treatment capacity and operating conditions of the target wastewater treatment processes are listed in Supplementary Table 1. The influent wastewater samples were collected before primary sedimentation except for that from WWTP E, where the influent wastewater was collected at the point of wastewater inflow into an aeration tank because influent wastewater directly flowed into the aeration tank without primary sedimentation. The sludge samples were collected from an aeration tank (or oxic tank). The treated effluent samples were collected after chlorination before being discharged to the environment. Samples were collected at each WWTP in dry weather three times both in winter (Dec 2018–Feb 2019) and summer (Jun–Aug 2019), at least two weeks apart. In February and August, samples were collected from WWTP B at each treatment stage (Fig. 1). All samples were transported to the laboratory on ice and processed within 24 h after sampling.
Sample processing
For the influent wastewater and sludge samples, 50 mL of the sample was first centrifuged at 10,000×g for 15 min. Next, DNA was extracted from the centrifuged pellets using a FastDNA Spin Kit for Soil (MPBiomedicals, USA). For the treated effluent samples, the samples were first concentrated via membrane filtration. The sample volume of the treated effluent was 200 mL, except that of the MBR effluent collected at WWTP B, where 2 L of the sample was filtrated. The solid particles in the treated effluent samples were collected on 0.2-µm polycarbonate membrane filter (K020G047A, Advantec-Toyo, Japan) through vacuum filtration. Thereafter, DNA was extracted directly from the membrane filter using the FastDNA Spin Kit for Soil (MPBiomedicals, USA). The concentration of each DNA extract was measured using a spectrophotometer (μCuvette Biophotometer, Eppendorf, Hamburg, Germany). A seasonal composite extract of DNA was prepared to eliminate daily fluctuations and obtain each seasonal average (winter or summer). To prepare the composite DNA extract, the extracts from the samples collected on the different three days (with at least 2-week interval) in the same season (winter or summer) for each sample type (wastewater, sludge, or treated effluent) at each WWTP were mixed together to achieve an equal mass (ng) of DNA. A total of 43 DNA extracts of the samples were prepared for the subsequent analyses, as listed in Supplementary Table 2.
Microbial population and community analysis based on 16 S rRNA gene
The microbial community of each sample was analyzed by targeting the V3-V4 regions of the 16 S rRNA gene. Accordingly, a sequence library was prepared via a two-step tailed polymerase chain reaction (PCR) using ExTaq Hot Start (Takara-bio, Japan). The primer sequences and PCR amplification conditions are listed in Supplementary Table 3. The prepared library was purified using a Wizard SV Gel and PCR Clean-UP System (Promega, Madison, USA). After validating the library quality using a Fragment Analyzer with a dsDNA 915 Reagent Kit (Agilent Technologies, USA), a paired-end sequence of 2 × 300 bp was acquired using a MiSeq System (Illumina, USA) with MiSeq Reagent Kit v3 (Illumina, USA). Raw sequencing data were trimmed and filtered using Fastx toolkit version 0.0.14 and sickle version 1.33. To obtain the paired-end library, trimmed reads were merged using FLASH version 1.2.11. Quantitative Insights into Microbial Ecology 2 (QIIME2) version 2020.2 was employed for taxonomy analysis. Chimeric sequences were removed using the dada2 plugin before classifying them into operational taxonomic units (OTUs) under the reference library of Greengene (ver. 13_8) with criteria of 97% identity. The sequence data were deposited in the DRA database of DNA Data Bank of Japan (DDBJ) with accession numbers of SAMD00281909–00281924, SAMD00281876–00281891, SAMD00206914–00206922, SAMD00233324–00233332, SAMD00282801 and SAMD00282798 (Supplementary Table 2).
The microbial population in each sample was quantified based on the 16 S rRNA gene using real-time PCR assay with a universal primer set of 341 F (5′-CCTACGGGNGGCWGCAG-3′) and 805 R (5′-GACTACHVGGGTATCTAATCC-3′)51. For the PCR assay, a PCR reaction mixture (20 μL) of each sample was prepared using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix Kit (Agilent Technologies, USA), after which amplification was detected using a real-time PCR system (Mx3000P, Agilent Technologies, USA). The PCR conditions were: denaturation at 95 °C for 3 min followed by 40 cycles of annealing at 55 °C for 45 s and extension at 72 °C for 1 min.
Shotgun metagenomic sequence analysis
The shotgun metagenomic sequences of the sample DNA extracts were then analyzed using a HiSeq X system. The DNA in a sample was fragmented to 500 bp using Covaris S220 (Covaris Inc., Massachusetts, USA). The sequence library was prepared using KAPA HyperPlus Kit with FastGene Adapter Kit (FastGene, Japan). After validation of the library quality using a Fragment Analyzer with a dsDNA 915 Reagent Kit (Agilent Technologies, USA), the paired-end sequence of 2 × 151 bp was acquired using a HiSeq X system (Illumina, USA). The raw paired-end sequences were filtered and trimmed using the Enveomics collection pipeline52. After quality trimming, the ARGs for each fragment sequence were identified using BLASTn against Comprehensive Antibiotic Resistance Database (CARD) ver. 3.0.753 with an E-value cutoff of 1 × 10−5. According to the BLAST output, the read count of each identified ARG was aggregated to obtain the “raw ARG profile,” which was composed of the list of ARGs and their read count present in each sample. The read count of a gene fragment tends to become larger when the original gene length is longer. To exclude the bias caused by the gene length, the read count of each ARG were normalized as reads per kilobase (RPK), according to the subject sequence length (slen) in the CARD database. The read count of the 16 S rRNA gene in each sample was counted using Parallel-META 354. The read count of 16 S rRNA gene was also normalized as RPK according to the 16 S rRNA gene length of E. coli as 1541 bp55. The normalized ARG profile (i.e., the list of ARGs and their RPK in a sample) was called “ARG profile.” The relative proportion of each ARG in a sample was calculated as the ratio of RPK of the ARG to the sum of RPK of all ARGs. The list of ARGs and their proportion in a sample was termed “ARG composition.” The total ARG abundance of a sample was calculated as the ratio of the sum of RPK of all ARGs to the RPK of 16 S rRNA gene. The shotgun metagenomic sequence data were deposited in the DRA database of DNA Data Bank of Japan (DDBJ) as accession numbers of SAMD00282059–00282074, SAMD00206783–00206785, SAMD00206872–00206884, SAMD00206896–00206913, and SAMD00282809–00282810 (Supplementary Table 2).
Multivariate analysis
The antimicrobial resistome and microbial community in the samples were compared using principal component analysis PCA via R version 4.0.0. The ARG composition data and genus-level OTU abundance data were used in the PCA to compare the antimicrobial resistome and microbial community, respectively. In both PCAs, scaling was performed to focus on the relative change in each component rather than the absolute change in abundant components.
Data availability
All sequence data are available from the DRA database of DNA Data Bank of Japan (DDBJ). The accession numbers of the sequence data are listed in the Methods and Supplementary Information. The original data and the high-resolution data table of Fig. 7 are available from Mendeley Data with the identifier https://doi.org/10.17632/n2fr5c32s9.1.
References
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
O’ Neil, J. Review on Antibiotic Resisitance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations (Health and Wealth Nations, 2014).
Rousham, E. K., Unicomb, L. & Islam, M. A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. B Biol. Sci. 285, 20180332 (2018).
Rosas, I. et al. Characterization of Escherichia coli isolates from an urban lake receiving water from a wastewater treatment plant in Mexico City: fecal pollution and antibiotic resistance. Curr. Microbiol. 71, 490–495 (2015).
Honda, R. et al. Impacts of urbanization on the prevalence of antibioticresistant Escherichia coli in the Chaophraya River and its tributaries. Water Sci. Technol. 73, 362–374 (2016).
Honda, R. et al. Estimated discharge of antibiotic-resistant bacteria from combined sewer overflows of urban sewage system. NPJ Clean Water 3, 15 (2020).
Kumar, M. et al. Prevalence of antibiotic resistance in the tropical rivers of Sri Lanka and India. Environ. Res. 188, 109765 (2020).
Xu, J. et al. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 119, 1379–1385 (2015).
Suzuki, S. & Hoa, P. T. P. Distribution of quinolones, sulfonamides, tetracyclines in aquatic environment and antibiotic resistance in Indochina. Front. Microbiol. 3, 67 (2012).
Guo, J., Li, J., Chen, H., Bond, P. L. & Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 123, 468–478 (2017).
Pazda, M., Kumirska, J., Stepnowski, P. & Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—a review. Sci. Total Environ. 697, 134023 (2019).
Zhang, T., Zhang, X.-X. & Ye, L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE 6, e26041 (2011).
Yang, Y., Li, B., Ju, F. & Zhang, T. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol. 47, 10197–10205 (2013).
Honda, R., Tachi, C., Noguchi, M., Yamamoto-Ikemoto, R. & Watanabe, T. Fate and seasonal change of Escherichia coli resistant to different antibiotic classes at each stage of conventional activated sludge process. J. Water Health 18, 879–889 (2020).
Tong, J. et al. Microbial community evolution and fate of antibiotic resistance genes along six different full-scale municipal wastewater treatment processes. Bioresour. Technol. 272, 489–500 (2019).
Threedeach, S., Chiemchaisri, W. & Chiemchaisri, C. Fate of antibiotic resistant E. coli in anoxic/aerobic membrane bioreactor treating municipal solid waste leachate. Int. Biodeterior. Biodegrad. 113, 57–65 (2016).
An, X. L. et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR. Environ. Int. 117, 146–153 (2018).
Chu, B. T. T. et al. Metagenomics reveals the impact of wastewater treatment plants on the dispersal of microorganisms and genes in aquatic sediments. Appl. Environ. Microbiol. 84, e02168–17 (2018).
Kotlarska, E., Łuczkiewicz, A., Pisowacka, M. & Burzyński, A. Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. 22, 2018–2030 (2015).
Port, J. A., Cullen, A. C., Wallace, J. C., Smith, M. N. & Faustman, E. M. Metagenomic frameworks for monitoring antibiotic resistance in aquatic environments. Environ. Health Perspect. 122, 222–228 (2014).
Kasuga, I., Nagasawa, K., Suzuki, M., Kurisu, F. & Furumai, H. High-throughput screening of antimicrobial resistance genes and their association with class 1 integrons in urban rivers in Japan. Front. Environ. Sci. 10, 1–15 (2022).
Rodríguez-Rojas, A., Rodríguez-Beltrán, J., Couce, A. & Blázquez, J. Antibiotics and antibiotic resistance: a bitter fight against evolution. Int. J. Med. Microbiol. 303, 293–297 (2013).
Chapman, J. S. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegrad. 51, 271–276 (2003).
Wales, A. & Davies, R. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604 (2015).
Lira, F., Vaz-Moreira, I., Tamames, J., Manaia, C. M. & Martínez, J. L. Metagenomic analysis of an urban resistome before and after wastewater treatment. Sci. Rep. 10, 1–9 (2020).
Ng, C. et al. Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Front. Microbiol. 10, 1–13 (2019).
Yang, Y., Li, B., Zou, S., Fang, H. H. P. & Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 62, 97–106 (2014).
Bengtsson-Palme, J. et al. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 572, 697–712 (2016).
Ping, Q. et al. The prevalence and removal of antibiotic resistance genes in full-scale wastewater treatment plants: bacterial host, influencing factors and correlation with nitrogen metabolic pathway. Sci. Total Environ. 827, 154154 (2022).
Zhang, S. et al. Fate of antibiotic resistant cultivable heterotrophic bacteria and antibiotic resistance genes in wastewater treatment processes. Chemosphere 135, 138–145 (2015).
Reinthaler, F. et al. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 37, 1685–1690 (2003).
Novo, A. & Manaia, C. M. Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 87, 1157–1166 (2010).
Ferreira da Silva, M., Vaz-Moreira, I., Gonzalez-Pajuelo, M., Nunes, O. C. & Manaia, C. M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 60, 166–176 (2007).
Stange, C., Sidhu, J. P. S., Toze, S. & Tiehm, A. Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int. J. Hyg. Environ. Health 222, 541–548 (2019).
Zhang, Y. et al. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 512–513, 125–132 (2015).
Yuan, Q.-B., Guo, M.-T. & Yang, J. Fate of antibiotic resistant bacteria and genes during wastewater chlorination: implication for antibiotic resistance control. PLoS ONE 10, e0119403 (2015).
Munir, M., Wong, K. & Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 45, 681–693 (2011).
Tchobanoglous, G. et al. Suspended growth bioological treatment processes. in Wastewater Engineering: Treatment and Resource Recovery (eds. Metcalf & Eddy, AECOM) 697–940 (McGraw-Hill Education, 2014).
Sulfikar, Honda, R., Noguchi, M., Yamamoto-Ikemoto, R. & Watanabe, T. Effect of sedimentation and aeration on antibiotic resistance induction in the activated sludge process. J. Water Environ. Technol. 16, 94–105 (2018).
Baker-Austin, C., Wright, M. S., Stepanauskas, R. & McArthur, J. V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182 (2006).
Aminov, R. I. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2, 1–19 (2011).
Jong, M., Harwood, C. R., Blackburn, A., Snape, J. R. & Graham, D. W. Impact of redox conditions on antibiotic resistance conjugative gene transfer frequency and plasmid fate in wastewater ecosystems. Environ. Sci. Technol. 54, 14984–14993 (2020).
Okubo, Y. et al. Change in clinical practice variations for antibiotic prescriptions across different pediatric clinics: a Japan’s nationwide observational study. J. Infect. Chemother. 27, 1621–1625 (2021).
Shishido, A. et al. Effects of a nudge-based antimicrobial stewardship program in a pediatric primary emergency medical center. Eur. J. Pediatr. 180, 1933–1940 (2021).
Sugawara, T. et al. Estimation of amount of antimicrobials used by pharmacy surveillance. Jpn. J. Infect. Prev. Control 27, 195–198 (2012).
Conco, T. et al. Profiling of emerging pathogens, antibiotic resistance genes and mobile genetic elements in different biological wastewater treatment plants. J. Environ. Chem. Eng. 10, 107596 (2022).
Calderón-Franco, D. et al. Metagenomic profiling and transfer dynamics of antibiotic resistance determinants in a full-scale granular sludge wastewater treatment plant. Water Res. 219, 118571 (2022).
Raza, S., Shin, H., Hur, H. G. & Unno, T. Higher abundance of core antimicrobial resistant genes in effluent from wastewater treatment plants. Water Res. 208, 117882 (2022).
Yuan, Q.-B. et al. Redistribution of intracellular and extracellular free and adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 131, 104986 (2019).
Wang, R., Matsuura, N., Hara-Yamamura, H., Watanabe, T. & Honda, R. Initial behaviors and removal of extracellular plasmid gene in membrane bioreactor. J. Environ. Manag. 298, 113541 (2021).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, 1–11 (2013).
Rodriguez-R, L. M. & Konstantinidis, K. T. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 4, e1900v1 (2016).
Jia, B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573 (2017).
Jing, G. et al. Parallel-META 3: comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci. Rep. 7, 40371 (2017).
Brosius, J., Palmer, M. L., Kennedy, P. J. & Noller, H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl Acad. Sci. USA 75, 4801–4805 (1978).
AMR Clinical Reference Center. Surveillance of antibiotic sales in Japan. https://amrcrc.ncgm.go.jp/surveillance/020/archives_en.html (2021).
Acknowledgements
This study was financially supported by JST MIRAI Program (Grant No. JPMJMI18DC), JSPS KAKENHI fund for Fostering Joint International Research (Grant Nos. 16KK0146, 18KK0114, 21KK0073), Grant-in-Aid for Scientific Research B (Grant No. 19H02272, 21H03617) and Kurita Water and Environment Foundation (Grant No. 22T007). Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Author information
Authors and Affiliations
Contributions
R.H.: conceptualization, methodology, formal analysis, writing—original draft, and supervision; N.M.: methodology, software, resource, and writing—review and editing; S.S.: investigation and data curation; S.A., Y. Morinaga, T.V.H.: investigation, data curation, and formal analysis; M.A.: data curation and writing—review and editing; Y. Masakke: methodology and software; H.H.-Y.: investigation, formal analysis, project administration, and writing—review & editing; T.W.: supervision, project administration, and funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Honda, R., Matsuura, N., Sorn, S. et al. Transition of antimicrobial resistome in wastewater treatment plants: impact of process configuration, geographical location and season. npj Clean Water 6, 46 (2023). https://doi.org/10.1038/s41545-023-00261-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41545-023-00261-x