Abstract
We designed and produced a self-assembling protein nanoparticle. This self-assembling protein nanoparticle contains five CD8+ HLA-A03-11 supertypes-restricted epitopes from antigens expressed during Toxoplasma gondii’s lifecycle, the universal CD4+ T cell epitope PADRE, and flagellin as a scaffold and TLR5 agonist. These CD8+ T cell epitopes were separated by N/KAAA spacers and optimized for proteasomal cleavage. Self-assembling protein nanoparticle adjuvanted with TLR4 ligand-emulsion GLA-SE were evaluated for their efficacy in inducing IFN-γ responses and protection of HLA-A*1101 transgenic mice against T. gondii. Immunization, using self-assembling protein nanoparticle-GLA-SE, activated CD8+ T cells to produce IFN-γ. Self-assembling protein nanoparticle-GLA-SE also protected HLA-A*1101 transgenic mice against subsequent challenge with Type II parasites. Hence, combining CD8+ T cell-eliciting peptides and PADRE into a multi-epitope protein that forms a nanoparticle, administered with GLA-SE, leads to efficient presentation by major histocompatibility complex Class I and II molecules. Furthermore, these results suggest that activation of TLR4 and TLR5 could be useful for development of vaccines that elicit T cells to prevent toxoplasmosis in humans.
Similar content being viewed by others
Introduction
Toxoplasma gondii infects all mammals. It can cause severe brain and eye damage in the fetus, in newborn infants, and in immune-compromised individuals.1 Although anti-parasitic medicines such as sulfadiazine and pyrimethamine are available, some patients experience side effects including toxicity and hypersensitivity. Latent, encysted parasites are not eliminated by these treatments.2 Therefore, development of a potent, safe, effective vaccine is greatly needed.
One approach for toxoplasmosis vaccine development is an epitope-based vaccine designed to enhance host immunity. Protection is achieved through stimulation of CD4+ helper T lymphocytes and CD8+ IFN-γ producing T lymphocyte responses. These CD8+ T cells recognize octamer/nonamer peptides presented on HLA supermotif molecules on infected cells. Previously, our laboratory (RM, KE) identified epitopes eliciting CD8+ T cells derived from proteins expressed during different phases of the Toxoplasma life cycle. HLA-A02, A03-11 and B07 human, supermotif, major histocompatibility complex (MHC) molecules are present in ~90% of humans,3,4,5,6 and therefore are capable of presenting these epitopes. As the discovery of such protective peptide epitopes accumulates, mechanisms are needed to effectively present these epitopes to the immune system of the host.
We have pioneered a platform known as Self-Assembling Protein Nanoparticles (SAPNs).7,8,9,10,11,12 SAPNs induce a strong immune response due to the repetitive display of antigens.7, 10, 12 They promote immune responses by CD4+ as well as CD8+ T cells by incorporating the T cell epitopes into the core architecture of the nanoparticle.8, 9, 11 They trigger a strong innate immune response by activating the TLR5 pathway through the adjuvant flagellin.13 Because of their size and shape they have the potential to reach follicular dendritic cells that are critical for antigen presentation and processing.14 Although macrophages play a role in immunity, interactions between SAPN and macrophages were not studied. SAPNs induce immune response that are orders of magnitudes stronger than Keyhole limpet hemocyanin, which is a standard vaccine carrier. We previously designed SAPN-based vaccine candidates for various infectious diseases including malaria,10, 11, 14, 15 HIV,16 SARS,17 and influenza.18
Earlier findings, and recent parallel work with a recombinant polypeptide, SAPNs, and GLA-SE (Fig. 1 and unpublished data [DL]) provide the foundation for our present studies. These earlier findings provide a basis for use of immunosense selected peptides from different genetic isolates of T. gondii (Fig. 1a), a flagellin scaffold,7, 8, 13, 19 and adjuvanting with GLA-SE.20,21,22,23 Earlier studies from the Walter Reed Army Institute of Research with malaria based SAPNs demonstrated that flagellin molecules improved immunogenicity (DL, PB, unpublished work). Initially, this was the basis for using flagellin as a SAPN scaffold in our T. gondii studies (Fig. 1b). This approach was also used in our work with influenza.24 This work suggested that flagellin would be helpful as a scaffold and immunogen in our newest T. gondii work.
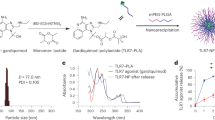
Assembly of CD8-SAPNs. a Phylogenetic tree showing 62 genetic isolates of Toxoplasma analyzed herein. These are in the multi-sequence alignments of proteins, and peptides derived from them, utilized to create our artificial immunogenic (“smart”) protein. b Flagellin is used as a scaffold into which epitopes are intercalated from Toxoplasma. Earlier logic for inclusion of flagellin as adjuvant and scaffold came from work with malaria (http://www.internationalinnovation.com/build/wp-content/uploads/2016/05/David_Lanar_Intl_Innovation_Infectious_Diseases_Research_Media_LR.pdf),19 as well as with influenza.24 Computer model of the prototype. The core particle composed of the pentameric and trimeric coiled coils is shown in green and blue, respectively. The following are attached to the trimeric coiled coil: the TRL5 agonist flagellin (D0 and D1 domains) (purple) with the A11 epitopes (yellow) and a CD4 epitope string (magenta). c SDS-PAGE of the purified protein. Lanes are as follows: Lane 1: MW (molecular weight markers). Lane 2–5: Elution fractions were from 19 to 22. Samples derive from the same experiment and the gels/blots were processed in parallel. d Transmission electron microscopy of the nanoparticle preparation. e SDS-PAGE 4–20% of the purified protein. Lane 1: MW (molecular weight markers). Lane 2: CD8-SAPN. Lane 3: Empty-SAPN. Samples derive from the same experiment and the gels/blots were processed in parallel
In experiments that provided a significant part of the foundation for our approach with SAPN to protect against toxoplasmosis, we (DL, PB, unpublished work) found the following: 1) GLA-SE or GLA-SE-like adjuvant was needed to produce significant titers of anti-nanoparticle antibody; 2) Purified IgG from immunized monkeys completely protected naïve mice (100%), when they were challenged with a lethal dose of 5000 sporozoites that express full-length Plasmodium. falciparum Circumsporozoite protein. Purified IgG from a control monkey did not protect any mice; 3) Purified IgG from immunized monkeys, mixed with P. falciparum sporozoites, prevented the sporozoite from infecting primary hepatocytes from human liver in tissue culture. IgG from control monkeys did not. Thus, we used this preliminary, foundational data when we chose GLA-SE as the adjuvant for our studies herein. GLA-SE has two components, GLA and SE. GLA is too hydrophobic to be used alone and any formulation of GLA would have other excipients making the formulation nonequivalent to GLA. Earlier studies demonstrated that the emulsion, called “SE”, did not adjuvant most proteins when administered alone. At present, GLA-SE is in pre-clinical studies or clinical trials as an adjuvant to prevent cancer, herpes, Leishmania, and Mycobacterium tuberculosis infections. Our earlier studies also demonstrated that GLA-SE was superior to ALUM as an adjuvant for our polypeptide.25 GLA-SE was also superior to ALUM in primates immunized with SAPN. In fact, ALUM diminished the response to GLA-SE plus SAPN (DL, PB, unpublished work).
In our previous studies with T. gondii, we constructed SAPNs displaying the dense granule epitope (GRA720–28) and pan-DR binding epitope PADRE.26 We evaluated these vaccine components in HLA-B*0702 transgenic mice.9 Immunization of these mice activated GRA7-specific CD8+ T cells that produced IFN-γ. Thereby, these mice were protected against subsequent challenge with high inocula of Type I and Type II parasites. These initial results highlighted the potential to protect against toxoplasmosis with a SAPNs vaccine approach.
In the present study, five epitopes which bind to HLA-A11-01 were evaluated for their efficacy in a SAPN-vaccine in HLA-A11-01 transgenic mice.5 These included epitopes from the surface antigen (SAG1), the dense granule proteins (GRA5 and GRA6), and the surface antigen-1-related sequences (SRS52A).5 In these constructs, the CD8+ HLA-A03-11 supertypes-restricted epitopes were linked by N/KAAA spacers. They were conjugated with PADRE, a universal CD4+ helper T lymphocyte epitope.26 This synthetic polypeptide is effective in mice and more effective than the pooled peptides separately.5, 23 PADRE binds promiscuously to MHC class II variants, and augments effector functions of CD8 + T cells through stimulation of IL2 production by CD4+ T helper cells.27, 28 Epitopes eliciting both CD4+ and CD8+ T cells are important components in the formulation of successful vaccines that drive protective responses.29 Our data show that incorporating PADRE into the SAPN constructs and delivering it in TLR4 ligand emulsion adjuvant (GLA-SE), resulted in activation of CD8+ T cells. This vaccine formulation led these cells to produce IFN-γ. They protected against subsequent challenge with Type II parasites given as a high inoculum. Thus, our work highlights the potential for the use of SAPN as a platform for the delivery of CD8+ and CD4+-restricted epitopes, formulated with the GLA-SE adjuvant, to protect against toxoplasmosis.
Results
Preparation and characterization of CD8+-SAPN and empty-SAPN
The SAPN constructs were expressed, purified and folded to form nanoparticles (Fig. 1c–e). The protein has a relative molecular weight of about 48 kDa on a Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1c, e). Transmission electron microscopy (Fig. 1d) showed a relatively uniform distribution of non-aggregated nanoparticles of about 30 nm in diameter.
In vivo immunogenicity of CD8+ T cell-eliciting SAPNs
HLA-A*1101-transgenic mice were immunized intramuscularly with CD8+ T cell-eliciting SAPNs combined with GLA-SE. Mice were immunized three times intramuscularly at 2 week intervals. Empty-SAPNs plus GLA-SE or PBS were used in sham immunizations of control mice. CD8+-T cell-eliciting SAPN-GLA-SE vs. Empty-SAPN-GLA-SE were compared in HLA-A*1101 transgenic mice as described. Spleen cells were obtained from immunized HLA-A*1101 transgenic mice 2 weeks after final immunization. IFN-γ produced by splenocytes cultured with the pool of peptides was measured. Figure 2 shows IFN-γ secretion is high in mice immunized with CD8+-T cell-eliciting SAPN plus GLA-SE when stimulated with PADRE or the GRA6 peptide. The other peptides also elicited IFN-γ production. In our earlier work,25 and herein, effects of the separate peptides were additive (Figs. 2b and 3). The polyepitopes elicited the best response earlier25 and herein (Fig. 3). Figure 3a and b indicate that IFN-γ secretion in cultures with the SAG1, GRA6, GRA3, and SRS52A peptides was significantly enhanced by immunization with these peptides but not Empty-SAPN or PBS. Significantly more IFN-γ secretion was observed when cells were stimulated with these pooled peptides plus PADRE. Thus, the association of CD8+ T cell- and CD4+ T cell-restricted peptides contributes to IFN-γ production in HLA-A*1101 transgenic mice.
CD8-SAPNs elicit restricted CD8+ T and CD4+ T cell peptide-specific immune response. ELISpot showing IFN-γ spot formation. Mouse splenocytes from Empty-SAPN, CD8-SAPN, and CD8-SAPN + GLA-SE were tested using GRA6 peptide (GRA6164–172) or PADRE. GLA designates GLA-SE in this figure. All peptides elicited IFN-γ (p < 0.05) compared to unstimulated cultures. Pooled peptides25 appeared additive. The greatest effect occurred with the polypeptide as also occurred in earlier studies.25 *p < 0.05
CD8-SAPNs are potent inducers of cell-mediated immunity. a IFN-γ ELISpot assay stimulated with a group of 5 peptides HLA-A*1101. b Graph shows the count of spots for splenocytes of untreated, Empty-SAPN + GLA1 CD8-SAPN + GLA-SE group of mice. GLA designates GLA-SE in this figure. *p < 0.05 for all IFN-γ ELISpots compared to controls
In vitro TLR5 stimulation
The SeaPorter TLR5 cell-line was exposed to varying concentrations of the SAPNs. The SAPN included: Empty-SAPN that do not contain the CD8+ epitopes but still have flagellin; CD8+-SAPN containing the polypeptide with the five restricted CD8+ epitopes; recombinant polypeptide; and recombinant flagellin (as control). The concentrations of SAPNs used were 0.01, 0.1, 1, 10, 100, and 1000 ng/ml. Fold increase in SEAP expression for each protein sample over non-treated controls reflected level of TLR5 stimulation. As shown in Fig. 4a–c, TLR5 activity was significantly enhanced by the Empty-SAPNs and the CD8+-T cell-eliciting SAPNs, but not the control polypeptide. Surprisingly, flagellin in Empty-SAPN particles have higher TLR5 activity than recombinant flagellin alone.
Seaporter TLR5 cell line responses to flagellin and SAPN. SeaPorter TLR5 cell-line was exposed to varying concentrations of each indicated protein, and the level of TLR5 stimulation was determined by the level of SEAP expression. Fold increase in SEAP expression for each protein sample over non-treated controls, error bars are standard error of the means. A two-way ANOVA model was fit with protein concentration and type as factors. There was a significant protein concentration by type interaction (p < 0.001); this indicated that the differences across types depended on the concentration and that the differences across concentrations varied by type. Specifically, there weren’t statistically significant differences across types at the two lowest concentrations (0.01 and 0.1), but there were significant differences between types at the 1, 10, 100, and 1000 ng/mL concentrations (p < 0.001 for all). Subsequent pairwise contrasts at these 4 concentrations found that the 5A11 Restricted CD8 + group (the recombinant protein without flagellin) was significantly different from the other 3 groups in all cases except for the 5A11 Restricted CD8 + vs. CD8-SAPN comparison at the 1 ng/mL concentration. In addition, at each of these 4 concentrations, the Empty-SAPN was significantly different (greater than) from the CD8-SAPN. There was a significant concentration effect for all protein types (p < 0.001) except 5A11 Restricted CD8 + (p > 0.9)
SAPNs with GLA-SE adjuvant confer robust protection against T. gondii in HLA-A*1101 transgenic mice
In the results shown in Fig. 5, we had immunized mice with either CD8+ T cell-eliciting SAPN with GLA-SE adjuvant, or Empty-SAPN with GLA-SE adjuvant, or adjuvant alone, or PADRE alone, or PBS. We then challenged 2 weeks after the last immunization with Type II strains of T. gondii expressing luciferase. Brains from these mice were imaged with a Xenogen camera 21 days after challenge with 2000 Me49-Fluc tachyzoites. Figure 5a and b show that luminescence from T. gondii in mice immunized with CD8+ T cell-eliciting SAPN plus GLA-SE was significantly lower than in mice immunized with control Empty-SAPN plus GLA-SE, GLA-SE alone, PADRE alone, or PBS. This finding correlates with a reduction of the number of cysts per brain in mice that received CD8+-T cell-eliciting SAPN plus GLA-SE adjuvant (Fig. 5c).
T. gondii brain cysts luciferase expression was significantly reduced in immunized HLA-A*1101 mice. HLA-A*1101 transgenic mice were immunized with GLA-SE adjuvanted Empty-SAPN or CD8-SAPN three times at intervals of 2 weeks. PBS was used as control. a T. gondii brain cysts luciferase expression was significantly reduced in HLA-A*1101 mice immunized with CD8-SAPN plus GLA boost at 21 days after challenge with 2000 Me49 (Fluc) T. gondii expressing luciferase. b Xenogen imaging of brain ex vivo following the injection of luciferin into the retro-orbital plexus and then exposure of the brain to luciferin solution. This figure shows data from mice in one of the replicate experiments (n = 4 control and 4 immunized mice). c Enumeration of cyst was performed with brains of mice challenged 21 days after final immunization. SAPN reduced cyst numbers and luminescence (p < 0.05)
Discussion
Improved vaccination and delivery approaches to elicit cellular immune responses against T. gondii are needed. In our previous studies we defined a panel of octamer/nonamer peptides restricted by MHC class I molecules. These peptide epitopes bind to and elicit IFN-γ responses from CD8+ T cells isolated from HLA A02, A03, and B07 individuals. These class I supermotifs are present in essentially all the human population worldwide, but with different frequency in different regions. When given with the GLA-SE adjuvant, these pooled peptides were able to protect haplotype specific HLA supermotif transgenic mice. This protection was measured as survival and reduced parasite burden.
Our capability to control the ability of peptides and proteins to self-assemble into particles which have a well-defined size and shape allows us to design mechanically and chemically stable particles. These SAPNs combine strong immunogenic effects of live attenuated vaccines with high specificity in eliciting immune responses of protein-based vaccines because they resemble virus capsids. It is apparent that the SAPNs have a great potential to serve as a platform for vaccines beyond their ability to present antigens in a repetitive manner. In contrast to live attenuated vaccines, SAPN-derived vaccines pose no significant risk of infection. They are very versatile and flexible in their design leading to better biophysical and immunologic properties. Furthermore, bacterial protein expression, purification, and self-assembly into nanoparticles reduces the time needed for large-scale vaccine production.
Herein, we used the SAPNs to present immunogenic peptide epitopes to a host’s immune system based on the assembly of five protective CD8+ CTL HLA-A03-11 restricted supertypes in addition to the universal helper epitope, PADRE. All epitopes were flanked at the C-terminus by N/KAAA spacers, which promote optimal immunogenic processing. Our data showed potent immunogenicity (high IFN-γ secretion) when splenocytes were stimulated by these peptides through immunization in vivo, and then exposure in vitro. In separate studies,24 we found that SAPN, which contains flagellin, protected better against influenza than SAPN without flagellin. This flagellin scaffold then became our SAPN platform going forward. In our TLR5 activity assay, the SAPN with the flagellin scaffold shows good stimulation of TLR5. However, the activity is reduced compared to the Empty-SAPN. This could be due to some interference with TLR5-binding and the presentation of the CD8+ T cell-restricted epitopes because the CD8+ epitopes string was engineered into the flagellin molecule to replace the D2 and D3 flagellin domains.
Thus, our future work will utilize this approach to engineer different SAPN constructs with optimized processing and immunogenicity for all our vaccine constituents. The proposed mechanisms for inducing innate immunity by our SAPN is the ligation of TLR4 by GLA in an emulsion30 and TLR5 by flagellin on the surface of the SAPN.13 McCoy et al.’s data suggested cross presentation14 of CD8+ stimulating epitopes in SAPNs (Fig. 1a). GLA-SE has been used with SAPN to successfully immunize against P. falciparum by eliciting antibody and T cells, whereas SAPN without GLA-SE was not effective (DL, PB, unpublished results). The adjuvant was safe in primates and now is entering clinical trials in humans. Despite remarkable protection provided by our SAPN vaccination in this study, some brain cysts were still detected. Thus, potential improvements in induction of protective immune responses could be made with the addition of separate nanoparticles with other CD4+ and CD8+ T cell-eliciting epitopes of various T. gondii proteins from several parasite life stages and potentially B-cell epitopes to stimulate a potent antibody response. Cell-mediated immunity, with cytolytic T cells and IFN-γ production, is considered to be the desired primary, protective, immune response.28, 31 Nonetheless, antibodies may contribute to protection. Addition of the micronemal proteins (MICs) or other proteins that induce antibodies that are neutralizing, adhesion or invasion blocking, or complement fixing, could further improve protection, if they play a significant part in attachment to or penetration of the host cell by the parasite. MICs have recently been used as recombinant vaccines and showed promising protection levels.32 MIC1 also stimulates IL 12 production in mice. Possibly, these proteins could also be engineered in separate SAPNs to yield a multi-SAPN vaccine to protect against toxoplasmosis.
Earlier studies provide support for using GLA-SE as an adjuvant for a wide variety of protein vaccines, including our own. We evaluated a P. falciparum SAPN vaccine and demonstrated GLA-SE was essential, or improved immunogenicity, vs. a related SAPN (DL, PB, unpublished). This study involved presenting antigens to protect against the phylogenetically related apicomplexan malaria parasite in non-human primates. It was found that GLA-SE was needed for immunization of primates even when this was not the case in mice. In other earlier work, separate constituents of GLA-SE were used. GLA alone was called “GLA-AF” when it was prepared in an aqueous formulation. This formulation did not contain an emulsion or extra excipients so it is not equivalent to the GLA in GLA-SE. GLA-AF, SE alone and GLA-SE formulations were compared in some of these earlier studies. In almost all of these different systems the value of using both GLA and SE together was proven (Table 1).20,21,22,23, 33,34,35,36 This work has been advanced to the clinic, demonstrating both efficacy and safety in studies with GLA and SE formulated and administered together. We leveraged this extensive, earlier experience to produce our GLA-SE adjuvanted SAPN vaccine for T. gondii.
In our work with this new SAPN-design, the flagellin molecule itself is an integral part of the SAPN scaffold (Fig. 1b), with or without the A11 CD8+ T cell-eliciting epitopes. This SAPN scaffold lacking the CD8+ epitopes conferred only a small amount of protection compared with the scaffold with the inclusion of the A11 peptides. It is not possible to create a relevant separate control without flagellin because in this scaffold the HLA A11 binding peptides are intercalated into the flagellin molecule itself as shown in Fig. 1.
The location of CD8+ epitopes within a protein sequence has been shown to be critical. Our arrangement of peptide epitopes into a polypeptide induced robust immunity.25 Deconvolution of peptide components has shown that certain epitopes alone may have different toxicity when separated from other peptides (El Bissati, McLeod, et al., in preparation). We already know that the adjuvanted polypeptide can protect in studies that are described in a separate manuscript.25 We also found that the HLA Class 1, A*1101 interacting peptides are specific for HLA A*1101 and not to other HLA supermotifs B7 or A2.25 Further, we demonstrated that the mouse C57Bl6 macrophages cannot present these peptides to HLA A*1101 T cells.25
Further, these five CD8+ epitopes, as well as full-length proteins from which they originate, were characterized to determine how well conserved the proteins, and especially the specific peptides we included, are across multiple strains of genetically divergent parasites from different geographic regions (Tables 2–4. Octamer/nonamer peptides; Supplementary figures S1 [SAG1], S2 [GRA6], S3 [GRA5], S4 [SAG2E], and S5 [SRS52A]). This analysis of 62 genetically divergent strains (Fig. 1a) supports the use of an immunosense approach. This approach creates a single protein which contains the relevant epitopes but does not include extraneous epitopes that are potentially harmful, as certain T. gondii epitopes are known to be. There were far fewer polymorphisms in our smaller octamer nonamer peptides (Tables 2–4) than in the full-length proteins (Table 3, Figs. S1–S5). There were good binding octamer/nonamers for three A11 epitopes among all the different genetic isolates (Table 4). The predicted binding scores for the peptides from the many genetically polymorphic strains were high for all but two peptides (Table 4). In contrast, it would take many full-length variants to obtain good geographic coverage of either of these polymorphic proteins if peptides other than our octamer/nonamer were critical to that protection. This provides further conceptual support for using an immunosense approach to make a vaccine with, potential to work well, and most parsimoniously, in many geographic areas. There are recent data concerning the unique processing of T. gondii proteins in human cells. Quite remarkably, there are longer (likely decoy) peptides that bind HLA-A2 in the natural infection of THP1 cells.37 This makes our targeted immunosense approach, beginning with human cells, then using HLA transgenic mice, especially valuable in creating a vaccine for people, rather than mice. These are critical considerations important beyond a vaccine protective against just this organism, when one wants to have broad utility across different demographics, worldwide. This is what we are working toward because of the substantial disease burden of toxoplasmosis as a global clinical problem.38
Data from studies of human cells and mice, in our work, considered together, demonstrate the robust and practical use of this model system.3,4,5,6, 25 Antigen processing and presentation in humans and mice have differences that are well known. For example, as shown elegantly, recently,39 human and murine tapasin diverge in sequence. These tapasins are chaperones of MHC class I molecules. Newer murine models with human tapasin and proteasome might improve vaccine potency and be more relevant to human vaccine development.39 Alternatively, our optimized cleavage sites, based on human infection, using human PBMC, may work as effectively in our HLA-A*1101 mice. This comparison will be tested in future studies. Further, there are other significant differences between mice and humans. Murine models have been imperfect predictors of vaccine performance in humans, apart from differences in HLA, tapasin or variants of ERAP. Differences in ERAP between mice and humans does not cause differences in processing and presentation. Other differences between mice and humans include very different total blood volume, rate of metabolism, skin composition/absorption rate, different TLR specificities, as well as other basic differences between murine and human cells. For example, human T cells, express Class II MHC, while mouse T cells do not. Nonetheless, relating to the question of whether HLA transgenic mice are a good approximation of human responses, Sette et al. made the following comment: despite the differences, when this issue was considered in detail, practical studies confirm that “although there ought to be a difference, actual data in peer reviewed studies, show that there is little difference”.40 Humanized mice with blood and liver transplants and other modifications are also still imperfect. These humanized mouse systems have not yet worked really well to demonstrate practical feasibility. For vaccine development, the FDA requires animal in vivo immunogenic data for IND submission. Vaccine developers have followed this requirement for the vaccines they are creating, which include viral vector or VLP-based vaccines. To date, there is no system that allows skipping the animal testing step. Most vaccines (e.g. viral, virus-like particles, proteins, bacterial) generate an immune response in an animal model. These immune responses provide preliminary data prior to studies in non-human primates and then in humans. This approach permits one to conclude that the vaccine is safe and ‘active’, even if the animal model is not absolutely predictive of precise human correlates, although that would be ideal. We have found that our approach appears to provide insight and to work effectively.3,4,5, 25 This approach includes using bioinformatics, testing human cells for immunogenicity, and then testing those down-selected peptides re-assembled into a protein with linkers designed for proper cleavage.41 This is followed by testing for efficacy and safety using HLA transgenic mice.3,4,5, 25 This approach is shown in our data herein, in our previous foundational experiments,25 and also by many others using other systems. This is for both immunogenicity of peptides or polypeptides or DNA or RNA in human cells first. This is then extended to murine cells, followed by protection measured as reduced parasite burden and enhanced survival. Although imperfect, there is considerable prior support, and support in these recent studies. The use of HLA transgenic mice can obviate problems of heterogeneity, both for MHC supermotifs, and parasite isolates. This is in a proven practical manner in vaccine development.19, 37, 42,43,44,45,46,47,48,49,50,51,52 The issues of genetic polymorphism, as well as potential harmful constituents, are amplified in full-length natural proteins (Tables 1–4; Fig. 1, S1–S5). Thus, the choice of peptides that are sufficient to interact with HLA molecules that are present in more than 90% of the human population can be made using bioinformatics in a rational and parsimonious manner. This approach considers parasite genetic variation in an inductive, immunosense manner that is proving to be valuable for development of vaccines for humans.
The inclusion of flagellin into immunogens can serve as a potential adjuvant.53-57 There is experience where flagellin has been safe and effective as an adjuvant in pre-clinical animal studies.53,54,55,56,57 It also has been effective when used for immunizations of both younger and older persons in clinical trials for influenza vaccines.53,54,55,56,57 There is a robust literature which describes studies of the mechanisms whereby this TLR5 ligand functions as an adjuvant.53-57
In summary, our study showed that a SAPN-protein chain with five CD8+ T cell-eliciting MHC class I epitopes from T. gondii, and the MHC class II epitope PADRE, can be refolded to form a nanoparticle. Using HLA-A*1101 transgenic mice, we demonstrate that the SAPN emulsified in GLA-SE adjuvant elicits a protective MHC class I response. Thus, our work demonstrates that we have developed an improved assembly of peptides for cross presentation of CD8+ T cell eliciting epitopes (Fig. 6) in vaccines to prevent toxoplasmosis.
SAPN adjuvanted with GLA-SE have peptides that are presented by MHC molecules on the follicular dendritic cells14 to T lymphocytes. GLA-SE and flagellin are ligands of TLR-4 and TLR-5 receptors, respectively. Ligating these receptors leads to the production of proinflammatory cytokines (IL-12, IL-6, TNF-α) and the expression of co-stimulatory molecules on the antigen-presenting cell surface. It remains to be determined whether the GLA-SE emulsion independently ligates TLR4 or whether SAPN are entrapped in the emulsion when this occurs, so both possibilities are shown. Original diagram for polyepitope for 5 A11 peptides25 provide a foundation to which concepts demonstrated in studies herein were added
Materials and methods
Peptides
KSFKDILPK (SAG1224–232), STFWPCLLR (SAG2C13–21), AVVSLLRLLK (GRA589–98), SSAYVFSVK (SRS52A250–258), AMLTAFFLR (GRA6164–172)25 and PADRE, a universal CD4+ helper epitope (AKFVAAWTLKAAA)26 were used in the vaccine constructs.25, 52 Infectious Diseases Research Institute (Seattle, Washington) synthesized the TLR4 agonist adjuvant called GLA-SE.3,4,5,6, 20,21,22,23, 25 This was prepared and used as a stable oil-in-water emulsion.
Molecular biology
The methods using DNA coding for the nanoparticle constructs were similar to those described in our earlier work.18 Briefly, they were prepared using standard molecular biology procedures as described in our earlier work from our laboratory by Babapoor et al.18 Specifically, plasmids containing the DNA coding for the protein sequence were used.18 They were constructed by cloning into restriction sites in the SAPN expression plasmid.18 We used a SAPN construct we had developed and described earlier.18 Briefly, this construct is composed of a pentameric coiled-coil tryptophan zipper.18 This zipper is linked by a glycine residue to a trimeric de-novo designed leucine zipper coiled coil.18 In this construct, a flagellin construct composed of the D0 and D1 domains (residues 1–177 and 249–372) of Salmonella enterica flagellin from the structure with pdb-code 3V47 from the RCSB protein data bank is used to extend the protein chain at the C-terminus18 (Fig. 1).
The CD8+-peptide sequence AVVSLLRLLKNAMLTAFFLRNAAAKSFKDILPKKAAASSAYVFSVKKAAAKFVAAWTLKAAAKSTFWPCLLR with the five CD8+ epitopes25 also containing PADRE25, 26 was next inserted into the D1 domain of flagellin.18 This polypeptide completely replaces the D2 and D3 domains to generate the CD8+ T cell-eliciting SAPN called “CD8-SAPN”.18 Overall, the positive charge of this epitope string is balanced with stretches of negative charges at both ends of the epitope sequence.18 Our Empty-SAPN was generated using the short linker KYKDGKGDDK to replace the D2 and D3 domains of flagellin.
Protein expression
This was performed exactly as we had performed and described in our earlier work from our laboratory by Babapoor et al.:18 Plasmids were transformed into Escherichia coli BL21 (DE3) cells.18 E. coli were grown at 37 °C in Luria broth with ampicillin.18 We induced expression using isopropyl β-D-thiogalacto-pyranoside. Cells were removed from 37 °C 4 h after induction.18 They were harvested by centrifugation at 4000 x g. We stored the cell pellet at −80 °C. We then thawed the cell pellet, keeping it on ice.18 We then suspended the pellet in a lysis buffer consisting of 9 M urea, 100 mM NaH2PO4, 10 mM Tris pH 8, 20 mM imidazole, and 0.2 mM Tris-2-carboxyethyl phosphine (TCEP). SDS-PAGE was used to assess our protein expression level.18
Protein purification
The same methodology we used earlier was used.18 Briefly, sonication was used to lyse cells, as described from our laboratory earlier.18 Centrifugation at 30,500 × g for 45 min18 was used to clarify the lysate. Then, for at least 1 h, our cleared lysate was incubated with Ni-NTA Agarose Beads (Qiagen, Valencia, CA, USA). Next, the column was washed with lysis buffer. This was followed by a wash with a buffer containing 9 M urea, 500 mM NaH2PO4, 10 mM Tris pH 8, 20 mM imidazole, and 0.2 mM TCEP. A pH gradient was used to purify the protein while bound to the column. The pH gradient for these wash steps was created as follows: 9 M urea, 100 mM NaH2PO4, 20 mM citrate, 20 mM imidazole, and 0.2 mM TCEP,18 with subsequent washes performed at pH 6.3, 5.9, and 4.5.18 To elute the protein, we used the lysis buffer, after the pH gradient, with a gradient of increasing imidazole concentrations.18
Protein refolding
We used methodology we have described in our earlier work.18 Specifically, for refolding, our protein was first rebuffered to the following conditions: 9 M urea, 20 mM Tris pH 8.5, 50 mM NaCl, 5% glycerol, 2 mM EDTA.18 4 µl of a solution with a concentration of 1.8 mg/ml protein was added to the same buffer solution without urea to a final concentration of 0.05 mg/ml for quick refolding of a first screen.18 We used this because this quick dilution from denaturing (urea) to native (no urea) buffer conditions triggers refolding of protein.18 We then used negative stain transmission electron microscopy at different resolutions to analyze our solution.18 Next, we used further screens for optimal refolding conditions.18 These were performed with smaller sampling sizes of the pH and ionic strength.18
In vitro TLR5 response assay
The methods were the same as those used in our recent work.28 Activation through TLR5 was assessed for SAPN as we described recently.24 Testing was done using TLR/NF-κB/SEAPorter™ Stably Transfected HEK 293 Cell Lines (Novus Biologicals, Littleton, CO; tested for Mycoplasma but not authenticated by STR profiling) as follows: All cell lines were stably co-transfected cell lines which express TLR5 and have a secreted alkaline phosphatase (SEAP) reporter gene under transcriptional control of an NF-κB response element. Fourteen thousand cells per well were seeded in a 96-well plate at passages 5–9. 20-4 h later, we removed growth media. Growth media was replaced with DMEM high glucose (Hyclone, Logan, UT). This contained either a SAPN, or recombinant flagellin (Novus Biologicals), at concentrations of 0.1, 1, 10, 100, 1000 ng/mL, each in triplicate. Media alone was present in control wells. Wells were exposed to the stimulus for 24 h. Then, supernatant was collected and used to determine whether SEAP was present. This was determined with a Reporter Assay kit for SEAP (Novus Biologicals). This was done using the manufacturer’s instructions. Media- only controls were used to normalize SEAP activity. This was used to determine each construct’s EC50. Triplicate determinations were utilized for each experimental condition.
Mice
The mice were those we created and described earlier.5, 23 The methods were identical to those used in our earlier work.3,4,5,6, 23 Specifically, “HLA-A*1101/Kb transgenic female mice were generated and then bred/produced at Pharmexa-Epimmune (San Diego, CA).23 They were then embryo-rederived at Taconic and JAX laboratories.23 Colonies were then expanded and they were then maintained and produced in isolators at the University of Chicago.23 These mice express a chimeric gene called HLA-A*1101/Kb transgene.23 This chimeric gene consists of the 1st and 2nd domains of HLA-A*1101 and the 3rd domain of H-2Kb.23 Mice were between 10 and 14 weeks of age in experiments. Mice were maintained in SPF conditions throughout.23 All of our studies were performed with the Institutional Animal Care and Use Committee at the University of Chicago’s review, approval, and oversight.
Immunizations of mice and challenge
To assess the immunogenicity of the SAPNs, mice with the HLA-A*1101 transgene were inoculated intramusculary. In this injection, 20 μg SAPN was emulsified in the TLR4 agonist, i.e., 5 μg of GLA-SE. The immunizations were administered three times at 2 weeks intervals. For the experiments in which these mice were challenged, challenge was at 14 days post-immunization.3 Specifically, they were challenged intraperitoneally using 2000 Type II (Me49-Fluc) parasites.23
ELISpot assay to determine murine splenocyte immune responses
This was performed as described in our earlier work which provided the foundation for our own present studies.3,4,5,6, 25 Specifically, spleens were harvested 14 days after immunization3,4,5,6, 25 as follows: initially, they were pressed through a 70 µm screen.3,4,5,6, 25 This allowed for formation of a suspension of single-cells. Erythrocytes were depleted from this suspension. AKC lysis buffer (160 mM NH4Cl, 10 mM KHCO3, 100 mM EDTA) was used to deplete the RBCs.3,4,5,6, 25 Hank’s Balanced Salt Solution (HBSS) was used to wash splenocytes twice.3,4,5,6, 25 Then the splenocytes were resuspended in RPMI-1640 supplemented with 2 mM L-GlutaMax.3,4,5,6, 25 Murine splenocyte ELISPOT assays were performed as described earlier.3,4,5,6, 25 This was done using anti-mouse IFN-γ mAb (AN18) and biotinylated anti-mouse IFN-γ mAb (R4–6A2).3,4,5,6, 25 In each well, 2.5–5 × 105 splenocytes were plated.3,4,5,6, 25 Mabtech (Cincinnati, OH) was the source of all of the antibodies and all of the reagents used to perform ELISPOT assays.3,4,5,6, 25 A minimum of three replicate wells were used to plate cells for each condition,3,4,5,6, 25 as described earlier, to measure spot-forming cells per 106 murine splenocytes.3,4,5,6, 25
Bioluminescence imaging to determinine outcomes of type II parasite challenge
We imaged mice infected with 2000 Fluc tachyzoites of the Me49 strain of T. gondii as described in our earlier work.25 Twenty-one days after challenge, an in vivo imaging system (IVIS; Xenogen, Alameda, CA)25 allowed us to visualize luciferin injected retroorbitally interacting with luciferase in the parasites.25 These mice were anesthetized. Anesthesia was performed in an O2-rich induction chamber with 2% isoflurane.25 Imaging took place 12 min after receiving luciferin.25 Living Image® 2.20.1 software (Xenogen) was used for assessment of photonic emissions.25 Pseudocolor representations of light intensity and mean photons/region of interest represent parasite burden in the imaging.25 All these mouse experiments were replicated a minimum of two times, as in our earlier work.25 In each group we used five mice.
Enumeration of cysts in mouse brains after type II parasite challenge
Mouse brains were collected at day 21, homogenized in 1 ml of saline (0.85% NaCl), and 50 µl of the homogenate was used to count the tissue cysts, microscopically, as described earlier.25 Cyst count was then multiplied by 20. This product then was used to determine the number of tissue cysts per brain.
Statistical analyses and additional detail concerning animal models
Data were compared for each assay by ANOVA and a Student’s t-test. GraphPad Prism 5 software (GraphPad Software, San Diego, CA) was used as described.6 ANOVA and multiple comparison procedures identified differences between the groups, as we previously described.6 Means ± SD are used to express data. A p value <0.05 was considered to be statistically significant for our results.6 Sample size in the in vivo studies was selected to be able to detect significant differences in luminescence based on our prior studies.25 With 5 per group, there is 80% power to detect a 2-standard deviation difference between groups. With 3 per group, there is 80% power to detect a 2.7-standard deviation difference between groups. All female mice we bred were utilized. They were randomly selected for the different groups but age-matched in the different groups within the experiment. There was no blinding in this experiment. In all in vivo experiments, there were 5 mice per group. In all in vitro experiments, there were 3 mice per group that provided splenocytes. All experiments were replicated at least twice. Representative experiments, of at least 2 separate trials, are shown. There was no data excluded from analyses.
Data availability
The data that support the findings of this study are available from ToxoDB (http://toxodb.org/toxo/) and the corresponding author on reasonable request.
References
McLeod, R., Kieffer, F., Sautter, M., Hosten, T. & Pelloux, H. Why prevent, diagnose and treat congenital toxoplasmosis? Mem. Inst. Oswaldo. Cruz. 104, 320–344 (2009).
McAuley, J. et al. Early and longitudinal evaluations of treated infants and children and untreated historical patients with congenital toxoplasmosis: the chicago collaborative treatment trial. Clin. Infect. Dis. 18, 38–72 (1994).
Tan, T. G. et al. Identification of T. gondii epitopes, adjuvants, and host genetic factors that influence protection of mice and humans. Vaccine 28, 3977–3989 (2010).
Cong, H. et al. Toxoplasma gondii HLA-B*0702-restricted GRA7(20-28) peptide with adjuvants and a universal helper T cell epitope elicits CD8(+) T cells producing interferon-gamma and reduces parasite burden in HLA-B*0702 mice. Hum. Immunol. 73, 1–10 (2012).
Cong, H. et al. Human immunome, bioinformatic analyses using HLA supermotifs and the parasite genome, binding assays, studies of human T cell responses, and immunization of HLA-A*1101 transgenic mice including novel adjuvants provide a foundation for HLA-A03 restricted CD8+ T cell epitope based, adjuvanted vaccine protective against Toxoplasma gondii. Immunome. Res. 6, 12 (2010).
Cong, H. et al. Towards an immunosense vaccine to prevent toxoplasmosis: protective Toxoplasma gondii epitopes restricted by HLA-A*0201. Vaccine 29, 754–762 (2011).
Burkhard, P. Peptidic nanoparticles as drug delivery and antigen display systems. US patent US8,575,110 B2. (2004).
Burkhard, P. Self-assembling peptide nanoparticles useful as vaccines. US patent US8,546,337 B2. (2009).
El Bissati, K. et al. Effectiveness of a novel immunogenic nanoparticle platform for Toxoplasma peptide vaccine in HLA transgenic mice. Vaccine 32, 3243–3248 (2014).
Kaba, S. A. et al. A nonadjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. J. Immunol. 183, 7268–7277 (2009).
Kaba, S. A. et al. Protective antibody and CD8+ T-cell responses to the Plasmodium falciparum circumsporozoite protein induced by a nanoparticle vaccine. PLoS ONE 7, e48304 (2012).
Raman, S., Machaidze, G., Lustig, A., Aebi, U. & Burkhard, P. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles. Nanomedicine 2, 95–102 (2006).
Raman, S. K. et al. Flagellin-containing protein nanoparticles as a vaccine platform. European patent application EP14,150,600 (2014).
McCoy, M. E. et al. Mechanisms of protective immune responses induced by the Plasmodium falciparum circumsporozoite protein-based, self-assembling protein nanoparticle vaccine. Malar. J. 12, 136 (2013).
Burkhard, P. & Lanar, D. E. Malaria vaccine based on self-assembling protein nanoparticles. Expert. Rev. Vaccines 14, 1525–1527 (2015).
Wahome, N. et al. Conformation-specific display of 4E10 and 2F5 epitopes on self-assembling protein nanoparticles as a potential HIV vaccine. Chem. Biol. Drug Des. 80, 349–357 (2012).
Pimentel, T. A. et al. Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem. Biol. Drug Des. 73, 53–61 (2009).
Babapoor, S. et al. A Novel Vaccine Using Nanoparticle Platform to Present Immunogenic M2e against Avian Influenza Infection. Influenza Res. Treat. 2011, 126794 (2011).
Lanar, D. E. The perfect vaccine, http://www.internationalinnovation.com/build/wp-content/uploads/2016/05/David_Lanar_Intl_Innovation_Infectious_Diseases_Research_Media_LR.pdf (2016).
Anderson, R. C. et al. Physicochemical characterization and biological activity of synthetic TLR4 agonist formulations. Colloids Surf. B. Biointerf. 75, 123–132 (2010).
Fox, C. B., Baldwin, S. L., Vedvick, T. S., Angov, E. & Reed, S. G. Effects on immunogenicity by formulations of emulsion-based adjuvants for malaria vaccines. Clin. Vaccine Immunol. 19, 1633–1640 (2012).
Baldwin, S. L. et al. Enhanced humoral and type 1 cellular immune responses with Fluzone adjuvanted with a synthetic TLR4 agonist formulated in an emulsion. Vaccine 27, 5956–5963 (2009).
Orr, M. T. et al. Adjuvant formulation structure and composition are critical for the development of an effective vaccine against tuberculosis. J. Control Release 172, 190–200 (2013).
Karch, C. P. et al. Vaccination with self-adjuvanted protein nanoparticles provides protection against lethal influenza challenge. Nanomedicine. doi:10.1016/j.nano.2016.08.030 (2016).
El Bissati, K. et al. Adjuvanted multi-epitope vaccines protect HLA-A*11:01 transgenic mice against Toxoplasma gondii. JCI Insight 1, e85955 (2016).
del Guercio, M. F. et al. Potent immunogenic short linear peptide constructs composed of B cell epitopes and Pan DR T helper epitopes (PADRE) for antibody responses in vivo. Vaccine 15, 441–448 (1997).
Brown, C. R. & McLeod, R. Class I MHC genes and CD8+ T cells determine cyst number in Toxoplasma gondii infection. J. Immunol. 145, 3438–3441 (1990).
Gazzinelli, R. T., Hakim, F. T., Hieny, S., Shearer, G. M. & Sher, A. Synergistic role of CD4+ and CD8+ T lymphocytes in IFN-gamma production and protective immunity induced by an attenuated Toxoplasma gondii vaccine. J. Immunol. 146, 286–292 (1991).
Alexander, J. et al. A decaepitope polypeptide primes for multiple CD8+ IFN-gamma and Th lymphocyte responses: evaluation of multiepitope polypeptides as a mode for vaccine delivery. J. Immunol. 168, 6189–6198 (2002).
van Haren, S. D. et al. In vitro cytokine induction by TLR-activating vaccine adjuvants in human blood varies by age and adjuvant. Cytokine 83, 99–109 (2016).
Suzuki, Y., Orellana, M. A., Schreiber, R. D. & Remington, J. S. Interferon-gamma: the major mediator of resistance against Toxoplasma gondii. Science 240, 516–518 (1988).
Pinzan, C. F. et al. Vaccination with recombinant microneme proteins confers protection against experimental toxoplasmosis in mice. PLoS ONE 10, e0143087 (2015).
Orr, M. T. et al. MyD88 and TRIF synergistic interaction is required for TH1-cell polarization with a synthetic TLR4 agonist adjuvant. Eur. J. Immunol. 43, 2398–2408 (2013).
Alving, C. R., Beck, Z., Matyas, G. R. & Rao, M. Liposomal adjuvants for human vaccines. Expert. Opin. Drug. Deliv. 13, 807–816 (2016).
Alving, C. R., Peachman, K. K., Rao, M. & Reed, S. G. Adjuvants for human vaccines. Curr. Opin. Immunol. 24, 310–315 (2012).
Alving, C. R., Rao, M., Steers, N. J., Matyas, G. R. & Mayorov, A. V. Liposomes containing lipid A: an effective, safe, generic adjuvant system for synthetic vaccines. Exp. Rev. Vaccines 11, 733–744 (2012).
McMurtrey, C. et al. Toxoplasma gondii peptide ligands open the gate of the HLA class I binding groove. Elife 5, doi:10.7554/eLife.12556 (2016).
McLeod, R. et al. Management of congenital toxoplasmosis. Curr. Pediatrics Rep. 2, 166–194 (2014).
Morozov, G. I. et al. Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc. Natl Acad. Sci. USA 113, E1006–1015 (2016).
Kotturi, M. F. et al. Of mice and humans: how good are HLA transgenic mice as a model of human immune responses? Immunome. Res. 5, 3 (2009).
Livingston, B. D. et al. Optimization of epitope processing enhances immunogenicity of multiepitope DNA vaccines. Vaccine 19, 4652–4660 (2001).
Paul, S., Sidney, J., Sette, A. & Peters, B. TepiTool: A Pipeline for Computational Prediction of T Cell Epitope Candidates. Curr. Protoc. Immunol. 114, 18, 19, 11–18 19 24 (2016).
Weiskopf, D. et al. HLA-DRB1 alleles are associated with different magnitudes of dengue virus-specific CD4+ T-cell responses. J. Infect. Dis. 214, 1117–1124 (2016).
Lindestam Arlehamn, C. S. et al. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis infected south africans. PLoS Pathog. 12, e1005760 (2016).
de Alwis, R. et al. Immunodominant dengue virus-specific CD8+ T Cell responses are associated with a memory PD-1+ Phenotype. J. Virol. 90, 4771–4779 (2016).
Bresciani, A. et al. T-cell recognition is shaped by epitope sequence conservation in the host proteome and microbiome. Immunology 148, 34–39 (2016).
Trolle, T. et al. The length distribution of class I-Restricted T cell epitopes is determined by both peptide supply and MHC allele-specific binding preference. J. Immunol. 196, 1480–1487 (2016).
Vita, R. et al. An ontology for major histocompatibility restriction. J Biomed Semantics 7, 1 (2016).
Sidney, J. et al. Characterization of the peptide binding specificity of the HLA class I alleles B*38:01 and B*39:06. Immunogenetics 68, 231–236 (2016).
Sette, A., Schenkelberg, T. R. & Koff, W. C. Deciphering the human antigenome. Exp. Rev. Vaccines 15, 167–171 (2016).
Carrasco Pro, S. et al. Automatic generation of validated specific epitope sets. J. Immunol. Res. 2015, 763461 (2015).
Alexander, J. et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity 1, 751–761 (1994).
Taylor, D. N., Treanor, J. J. & Strout, C. et al. Induction of a potent immune response in the elderly using the TLR-5 agonist, flagellin, with a recombinant hemagglutinin influenza-flagellin fusion vaccine (VAX125, STF2.HA1 SI). Vaccine 29, 4897–4902 (2011).
Taylor, D. N., Treanor, J. J. & Sheldon, E. A. et al. Development of VAX128, a recombinant hemagglutinin (HA) influenza-flagellin fusion vaccine with improved safety and immune response. Vaccine 30, 5761–5769 (2012).
Turley, C. B., Rupp, R. E. & Johnson, C. et al. Safety and immunogenicity of a recombinant M2e-flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine 29, 5145–5152 (2011).
Treanor, J. J., Taylor, D. N. & Tussey, L. et al. Safety and immunogenicity of a recombinant hemagglutinin influenza-flagellin fusion vaccine (VAX125) in healthy young adults. Vaccine 28, 8268–8274 (2010).
Schijns, V. E. J. C. & O’Hagan, D. T. Immunopotentiators in Modern Vaccines. (Elsevier Academic Press, 2016).
Acknowledgements
We thank and are grateful to Nihlab Shastri (University of California, Berkeley) and Lo Vang (PaxVax) for critical advice and suggestions. We gratefully acknowledge the Mann-Cornwell, Morel, Engel, Rooney–Alden, Pritzker, Langel, Drago, and Rosenthal families for their support of this work. This project has been funded in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under Award Numbers DMID-NIAID U01 AI77887 (to RM), R01 AI027530 (to RM), and U19 AI110819 (to HL). The research was also supported by the Knights Templar Eye Foundation (to KE) and the Research to Prevent Blindness Foundation (to RM).
Author information
Authors and Affiliations
Contributions
K.E., P.B., S.R., D.E.L., H.L., and R.M. designed research; K.E., Y.Z., I.J.B., S.P., S.K.R., J.S., and C.K. performed research; K.E., Y.Z., S.P., S.K.R., C.K., C.R., D.E.L., S.R., C.F., D.C., J.A., A.S., H.L., P.B., and R.M analyzed data; and K.E., P.B., and R.M. wrote the paper. All authors read and approved the final manuscript version.
Corresponding authors
Ethics declarations
Competing interests
P.B. has an interest in Alpha-O Peptides, a company with a focus on SAPN. R.M. agreed to be on the scientific advisory board for this company. This company has patents or patents pending on relevant technology. The following authors (R.M., K.E.B., Y.Z., P.B., J.A., A.S., J.S., S.R., C.F., D.C., C.W.R.) have submitted a patent application covering much of our vaccine work over the past years, including the work described in this manuscript. This is with the ultimate goal of making a vaccine that can be moved into the clinic to prevent suffering from human toxoplasmosis, so that this vaccine can be widely available for humans. The other authors declare no competing interests. Funding sources had no influence in study design; nor in the collection, analysis and interpretation of data; nor in the writing of this report; and no interest in the decision to submit the paper for publication.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El Bissati, K., Zhou, Y., Paulillo, S.M. et al. Protein nanovaccine confers robust immunity against Toxoplasma . npj Vaccines 2, 24 (2017). https://doi.org/10.1038/s41541-017-0024-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41541-017-0024-6
This article is cited by
-
Toxoplasmosis vaccines: what we have and where to go?
npj Vaccines (2022)
-
From Bench to the Clinic: The Path to Translation of Nanotechnology-Enabled mRNA SARS-CoV-2 Vaccines
Nano-Micro Letters (2022)
-
PLGA Nanoparticles as an Efficient Platform in Protein Vaccines Against Toxoplasma gondii
Acta Parasitologica (2022)
-
Building Programs to Eradicate Toxoplasmosis Part IV: Understanding and Development of Public Health Strategies and Advances “Take a Village”
Current Pediatrics Reports (2022)
-
Building Programs to Eradicate Toxoplasmosis Part I: Introduction and Overview
Current Pediatrics Reports (2022)