Abstract
Rheumatoid arthritis (RA) seriously impairs the quality of life of sufferers. It has been shown that Lycium barbarum polysaccharide (LBP), a natural active indigestible ingredient with medicinal and edible functions, can effectively relieve RA, however, whether this effect is related to gut microbiota is not known. This study aimed to explore the RA alleviating mechanism of LBP mediated by gut microbiota using a collagen-induced arthritis rat model. The results showed that LBP significantly changed the gut microflora structure accompanied with the RA alleviation. Specifically, a LBP intervention reduced the relative abundance of Lachnospiraceae_NK4A136_group and uncultured_bacterium_f_Ruminococcaceae and significantly increased the abundance of Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum. The mRNA contents of several colonic epithelial genes including Dpep3, Gstm6, Slc27a2, Col11a2, Sycp2, SNORA22, Tnni1, Gpnmb, Mypn and Acsl6, which are potentially associated to RA, were down-regulated due to the DNA hypermethylation, possibly caused by the elevating content of a bacterial metabolite S-adenosyl methionine (SAM). In conclusion, our current study suggests that LBP alleviated RA by reshaping the composition of intestinal microflora which may generate SAM, inducing DNA hypermethylation of RA-related genes in the host intestinal epithelium and subsequently reducing their expression.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease of unknown etiology, and characterized by infiltration of activated immune cells and production of inflammatory factors leading to synovial hyperplasia and pannus, as well as the destruction of cartilage and joints. Clinically, RA manifested as joint swell and stiffness, musculoskeletal pain and irreversible bone damage, and even permanent disability, which can seriously affect the quality of life1,2,3. At present, due to the potential side effects of drugs on the clinical therapy of RA, researchers are actively exploring other treatments. The development of active substances from natural plants has become a research hotspot.
Lycium barbarum has a great edible and medicinal value. As one of the most important bioactive ingredients in Lycium barbarum, Lycium barbarum polysaccharide (LBP) has multiple biological functions and pharmacological effects, such as anti-oxidation4, strengthening the barrier5, anti-tumor6, anti-inflammatory7, and regulating intestinal flora8. Recently, LBP was reported to effectively improve the clinical condition of RA by maintaining the bone integrity of type-II collagen-induced arthritis (CIA) rats and reducing the CIA-stimulated inflammatory mediators9. LBP can also affect the proliferation of chondrocytes and reduced arthritis10. Therefore, LBP is clearly a potential active compounds for RA treatment with low toxicity and low side effects. However, due to the complicated pathogenesis of RA, the mechanism of LBP on RA relief remains unclear.
LBP belongs to the non-starch polysaccharides (NSPs), which cannot be directly digested and absorbed by the body. NSPs will directly enter the large intestine, where they will be broken down by intestinal flora. More and more evidences prove that RA is closely related intestinal microecology11. Literatures showed enteric malnutrition and intestinal flora imbalance were typical features of RA12, while restoring intestinal homeostasis was proposed to be a therapeutic strategy to reduce RA13.
Intestinal microflora participated in the epigenetic shaping of host genes14, and modulated gene expression in intestinal epithelium15. But it is still elusive whether LBP modulated the gut microbiota, and how these gut bacteria played a role in RA alleviation. The aim of this study is to explore the potential mechanism of RA alleviation by LBP from the aspects of intestinal flora reshaping and intestinal epithelium gene regulation.
Results
RA-caused paw swell and joint pathological change were relieved by LBP
Paw swell and arthritis score were used to assess the severity of arthritis. Paws of rats in the Model group were swollen during the whole experiment (Fig. 1a) as indicated by the significant larger diameter of paws (Fig. 1b) and higher arthritis score (Fig. 1c) comparing with those in the Control group (P < 0.01). Joint structure of rats in the Model group was abnormal, along with the infiltration of inflammatory cell, synovial and fibrous tissue hyperplasia, and the occurrence of synovial hypertrophy and pannus (Fig. 1d).
a Joint swell of the left hind foot. b The diameter of paw swell in each group (mm). c The arthritis score of rats in each group. The error bars on the bar charts represented the standard deviations. The “*” indicated the P value was less than 0.05, “**” indicated the P value was less than 0.01, and “***” indicated the P value was less than 0.001. d Pathological sections of joint after HE staining in each group. Black arrow: diffusion or infiltration of inflammatory cells. Red circle: synovial hyperplasia and hypertrophy and formation of pannus. The magnification was 100×. The length of the scale bar was 250 μm.
The administration of LBP relieved the severe arthritis (Fig. 1a). Comparing with the Model group, the average diameter of paw in the Lyc group significantly reduced (Fig. 1b) implying the paw swell was greatly improved. Similar result was also obtained from the arthritis score (Fig. 1c). In addition, rats in the Lyc group had less inflammatory infiltration, and no large areas of synovial hyperplasia, and did not form pannus as shown in hematoxylin and eosin (HE) staining (Fig. 1d).
LBP reduced the content of pro-inflammatory and recovered the content of anti-inflammatory cytokines
Pro-inflammatory factors including IL-1α, IL-1β, IL-12 and IL-17 abnormally increased while anti-inflammatory factor abnormally decreased in RA. Among them, both IL-1 and IL-17 are considered as important indicators in the pathogenesis of RA. They can aggravate autoimmune pro-inflammatory responses, promote synovial hyperplasia, and lead to cartilage erosion16. In our experiments, the contents of pro-inflammatory cytokines IL-1α, IL-1β, IL-12 and IL-17 in the Model group were significantly higher than those in the Control group (Fig. 2a–d), while the anti-inflammatory cytokine IL-10 was on the contrary (Fig. 2e). After the administration of LBP, the contents of serum IL-1α, IL-1β, IL-12 and IL-17 in the Lyc group significantly reduced (Fig. 2a–d), while the content of IL-10 was recovered and tend to become normal (P < 0.05) (Fig. 2e). Together with the results of the pathological changes, LBP could significantly alleviated RA.
LBP altered specific intestinal bacteria that correlated to the inflammatory cytokine level and RA symptoms
On the whole, a total of 1,840,370 valid reads were obtained from all samples. The intestinal microflora on the Operational Taxonomic Unit (OTU) level showed that all groups shared the vast majority of OTU, but it’s worth noting that 6 unique OTUs in the Lyc group (Fig. 3a). Moreover, the intervention of LBP reconstructed intestinal microflora as indicated by the apparent distance among different groups (Fig. 3b). On the phylum level (Fig. 3c), the relative abundance of Fusobacteria was higher in the Model group than that in the Lyc group, which means the LBP intervention significantly altered the growth of Fusobacteria (Table 1). As for the genus level, when compared with the Model group, the Lyc group showed a lower relative abundance of Lachnospiraceae_NK4A136_group and uncultured_bacterium_ f_Ruminococcaceae, and a higher relative abundance of specific bacteria, including Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum (Fig. 3d). The altering of microbial compositions manifested the LBP significantly reconstructed the intestinal microflora in RA rats.
Correlation analysis revealed that both Lachnospiraceae_NK4A136_group and uncultured_bacterium_f_Ruminococcaceae were positively correlated to the content of pro-inflammatory cytokines, paw swelling and arthritis scores while negatively correlated to the content of IL-10. In contrast, Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum were negatively correlated to those factors except the IL-10 (Fig. 4). These results indicated elevating abundance of Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum by LBP intervention played a role in the inflammation reduction and the RA alleviation.
LBP altered the transcriptome of colonic epithelial tissue
In total, 462 genes were differentially up-regulated and 293 were down-regulated in the Lyc group when compared with those in the Model group (Fig. 5a). These differentially expressed genes (DEGs) can be classified into multiple KEGG classes (Fig. 5b, top 30 classifications in level 2 were shown). Among all classifications, the immune system covered the largest number of DEGs, followed by global and overview maps and signal transduction. Further evaluation for the enrichment analysis showed 32 KEGG pathways were significantly enriched (Fig. 5c). Nearly half of the pathways (15/32) belonged to the classification of ‘Human Diseases’, and 14/15 of these pathways related to infectious or immune disease. In addition, some inflammation-related pathways such as TNF signaling pathway, PPAR signaling pathway and cytokine-cytokine receptor interaction were also included. The transcriptomic results implied the intervention of LBP may have some impacts on immune-related genes in colonic epithelial tissue.
a Volcano plot of differentially expressed genes (DEGs) between groups. b The top 30 KEGG classes which DEGs were classified. Noted that, CP: Cellular Processes, EIP: Environmental Information Processing, GIP: Genetic Information Processing, OS: Organismal Systems. c The KEGG pathways which DEGs significantly enriched. d Distribution of differentially methylated regions (DMR) and e differentially methylation sites (DMS) on different gene elements. Noted that, different colors and numbers in circle represented different gene elements.
LBP reprogramed DNA methylome, and thereby regulated the expression of specific genes to alleviate RA
According to the results of methylome, 697 differentially methylated regions (DMRs) annotating 375 genes were found between the Lyc and Model groups. Among them, 6.9% located in the promoter region (Fig. 5d). Additionally, 51,298 nucleotide sites were methylation-specific modified, and most of them are located in intergenic and intron, while 5.6% and 8.1% are located in promoter and exon regions, respectively (Fig. 5e). Hypermethylation, especially for that occurred in the promoter region, is thought to inhibit gene expression by preventing the transcription factors from combining with the gene17,18. Together with the results of transcriptome, 89 genes in the Lyc group were found to be in high methylation level and simultaneously showed suppressed expression when using the Model group as the control. We further screened out 10 genes including Dpep3, Gstm6, Slc27a2, Col11a2, Sycp2, SNORA22, Tnni1, Gpnmb, Mypn and Acsl6 based on the number of methylation site and/or the region of methylation (Table 2). All these genes were found to have a higher fragments per kilobase of exon per million mapped reads (FPKM) value but a lower methylation level in the Model group while they have a lower FPKM value but a higher methylation level in the Lyc group.
The expression of these genes was positively correlated to either the content of certain pro-inflammatory cytokines or the paw swell and arthritis score with statistically significance (Fig. 4). Specifically, the correlation between the gene expression of Dpep3, Mypn and the content of IL-1α, IL-1β, IL-17A; Col11a2, Gpnmb and IL-1β, IL-17A; SNORA22 and IL-1α, IL-12; Acsl6 and IL-1β, IL-12; Gstm6 and IL-1α; Slc27a2 and IL-1α; Tnni1 and IL-1β was significantly positive (P < 0.05). Additionally, the expression of Acsl6 was negatively correlated to the content of IL-10 with significance (P < 0.05). Furthermore, the expression of Tnnil1, Gpnmb, Mypn and Acsl6 was positively correlated to the paw diameter (paw swell) while the expression of SNORA22 and Acsl6 was positively correlated to the arthritis score (P < 0.05). Therefore, the downregulation of these genes was closely connected to the inflammation and RA alleviation.
The methylome reprograming was modulated through direct methyl donor SAM potentially generated by LBP-intervened gut microbiota
Previous studies have shown that intestinal microorganisms affect the expression of host genes by impacting the epigenetic modification of host genes19,20. Therefore, we speculate that the suppressed expression of those specific genes mentioned above due to the DNA hypermethylation could be related to the gut microbiota intervened by LBP. Based on the results of the correlation analysis, the mRNA content of Acsl6 was negatively correlated to the relative abundances of Romboutsia and Lactobacillus while the mRNA contents of both Slc27a2 and SNORA22 were negatively correlated to the abundances of all four bacteria (Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum) with significance. The mRNA contents of other genes were also negatively correlated to the abundances of Romboutsia, Dubosiella and Faecalibaculum (P < 0.05) (Fig. 4).
Further identification of the substance accounted for hypermethylation was applied. The results showed that the content of SAM in the colonic epithelium in the Lyc group significantly increased compared with that in the Model group (Fig. 6a) (P < 0.05). Judging from the correlation analysis, we found a strong positive correlation between the content of epithelial SAM and the relative abundance of 4 specific gut bacteria (Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum) (Fig. 4). In addition, the content of SAM inside the intestinal contents (bacterial SAM) significantly was higher in the Lyc group than the Model group (Fig. 6b) (P < 0.05). These results may show a possibility that the additional production of SAM (the direct methyl donor possibly caused hypermethylation in specific genes) in the Lyc group was due to the increase of those 4 bacteria.
Discussion
RA is a chronic disease ubiquitous all over the world. It is the result of complex interactions among genes, environmental and hormonal factors and the immune system21. In our experiment, LBP relieved the typical features of RA in rats, including the reduction of paw swell, arthritis scores and the contents of pro-inflammatory cytokines such as IL-1α, IL-1β, IL-12 and IL-17, and the recovery of the content of anti-inflammatory cytokine IL-10. In addition, LBP improved the pathological manifestations of joint and cartilage in RA rats. Multi-omics analysis also manifested that LBP reshaped the gut microbial structure, specifically the modulation of the relative abundance of Romboutsia, Lactobacillus, Dubosiella, Faecalibaculum, Lachnospiraceae_NK4A136_group and uncultured_bacterium_f_Ruminococcaceae, and regulated the gene expression in colonic epithelium, especially for the suppressed expression of Dpep3, Gstm6, Slc27a2, Col11a2, Sycp2, SNORA22, Tnni1, Gpnmb, Mypn and Acsl6 due to the DNA hypermethylation.
It is widely accepted that intestinal microflora is a factor affecting metabolic homeostasis and the immune system22. The researcher has proposed the concept of the gut-joint axis, implying connections between intestinal microflora and RA. For instance, in the early stages of RA, the number of Bifidobacteria and Bacteroidetes decreased while the number of Prevotella increased23. It is also reported that the gut microbiome in the new-onset RA was characterized by an increase of P. Copri and a reduction of Bifidobacteria24. Therefore, the alleviation of RA by LBP should be attributed to its capability for gut microbiota modulation. In this current study, we found specific bacteria including Lachnospiraceae_NK4A136_group, uncultured_ bacterium_f_Rumino-coccaceae, Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum were regulated. Among these bacteria, Fusobacteria and Lachnospiraceae are reported to associate with inflammatory bowel disease25,26,27, while inflammatory bowel disease is considered as the predisposing factor of RA28. It seems that LBP could alleviate RA by reducing some susceptibility factors caused by gut microbiota. Additionally, LBP recovered the abundance of Romboutsia and Faecalibacterium, whose loss could lead to intestinal inflammation and incompleteness of intestinal epithelium. In this regard, LBP might facilitate the colonization of beneficial microorganisms in the intestine, enhance the intestinal barrier function, relieve the intestinal permeability, maintain intestinal integrity and intestinal health, and promote intestinal development29. LBP intervention also elevated the abundance of Lactobacillus, which is a characteristic of bacteria in rheumatoid joint diseases. It is reported that joint swelling and bone destruction can be improved by increasing the abundance of Lactobacillus30. Particularly, L. salivarius or L. plantarum showed RA amelioration from clinical manifestations31, and L. fermentum as well as L. swissericus also alleviated RA32, and even showed a certain ability for RA prevention33. Hence, the LBP-intervened microbiota should play an important role in the mechanism of RA alleviation.
The alteration of microbial structure caused by NSPs’ consumption was commonly accompanied with the change of gene expression in the intestinal epithelium. As predicted, the expression of over 700 genes were differentially regulated. We further found over 60% of these DEGs were methylated modified. Methylation modification is one of the epigenetic changes which altered the DNA configuration but not the nucleotide sequence to affect the expression of genetic material34. Gene expression is negatively correlated with the degree of methylation, and genes in hypermethylation are transcriptionally suppressed35. Since LBP lessened the RA, we focused on those down-regulated genes caused by DNA hypermethylation and eventually screened out 10 genes including Dpep3, Gstm6, Slc27a2, Col11a2, Sycp2, SNORA22, Tnni1, Gpnmb, Mypn and Acsl6, which can be divided into 3 categories.
Genes related to the alleviation of RA and RA-associated lesions including Col11a2, Acsl6, Gpnmb and SNORA22, can be classified into category A. Among them, Col11a2 is a gene encoding type XI collagen, which plays a key role in maintaining the characteristics of the cartilage matrix36. Studies have shown that the expression of Col11a2 was significantly increased in the cartilage of patients with arthritis, the overexpression of Col11a2 promoted the destruction of articular cartilage and chondrocyte necrosis37. LBP reduced the expression of Col11a2, which slowed down the degradation of articular cartilage and relieved RA. In addition, the overexpression of Acsl6 in chondrocytes induced the typical characteristics of arthritis, leading to the down-regulation of type-II collagen and the destruction of articular cartilage38. Since LBP had caused the high methylation and low expression of Acsl6 gene, it should inhibit the destruction of articular cartilage, and thus alleviated arthritis. Angiogenesis was reported to positively correlate with the severity of RA39. As a gene highly expressed in tumors40, Gpnmb promoted angiogenesis41,42. Thus, its suppressed expression after LBP consumption may reduce the angiogenesis and further alleviate RA. The overexpression of SNORA22 positively correlated with arthritis because it contributes to cell invasion and tumor metastasis, while cell erosion can aggravate the disease manifestations of RA43. In our experiment, LBP inhibited SNORA22 gene expression by elevating the methylation level, and may eventually improve RA.
Genes related to the inflammation including Slc27a2, Gpnmb, Gstm6 and Acsl6, can be classified into category B. Obesity can be considered as a chronic inflammation with low grade, and Slc27a2 was reported to be a potential marker of obesity44. Its expression was positively correlated to both content of IL-6 and TNF-α in peripheral blood mononuclear cells isolated from obese human45. Gpnmb was highly expressed in macrophage, and its content was increased in pro-inflammatory conditions46. It promoted the production of TNF-α in in vitro cell test47. The LBP intervention in the current research lowered the expression of both Slc27a2 and Gpnmb by remodeling their DNA methylome, and subsequently relieved the inflammation. Gstm6 and Acsl6, were also regarded as the candidate genes related to inflammation in hepatic lesions such as hepatitis and liver cancer48,49. However, interestingly, the low expression of these 2 genes enhanced the inflammation were reported, which was different from our results. This is probably due to different disease models applied in those reports. But further study will be needed to clarify the exact function of these 2 genes in RA.
Similarly, the rest of genes including Dpep3, Sycp2, Tnni1 and Mypn, which can be classified into category C, should also be addressed since no literatures reported their connections to RA or inflammation. Nevertheless, based on our current study, we believed they also played an important role in RA alleviation.
Taken together, the suppressed expression of these 10 epithelial genes due to their hypermethylation by LBP intervention may lower the content of proteins that associated with RA-caused lesions or inflammatory cytokines, and their transportation towards joint through blood flows were then possibly reduced. Therefore, LBP intervention may alleviate RA through gut-joint axis.
DNA methylation modulated the gene expression. As the direct methyl donor, SAM is critical in the process of DNA methylation. It transfers its own active methyl groups to specific bases in the DNA strand to methylate DNA and directly affects the degree of DNA methylation. Our results demonstrated LBP consumption recovered the insufficient SAM caused by RA, ultimately leading to DNA hypermethylation and gene hypo-expression. It should be noted that, SAM is also a common and important metabolite of intestinal microflora. Studies have confirmed that SAM from gut bacterial fermentation could be available to the host, and it is a significant, yet underestimated source of SAM for the host50. As one type of the NSPs, LBP reconstructed the gut microbiota, especially for the elevation of Lactobacillus, and could be subsequently metabolized into SAM. This was supported by some documents reporting that SAM was produced by in vitro fermentation of Lactobacillus51. In addition, Lactobacillus was listed as excellent SAM-producing bacteria among multiple SAM-producing probiotics50,51. Therefore, the hypermethylation of specific genes in intestinal epithelium should be the result of the SAM content recovery. This recovery possibly owing to the alteration of gut microbiota, especially for the increased abundance of several gut bacteria such as Lactobacillus after LBP intervention.
Collectively, the intervention of LBP relieved RA effectively. The underlying mechanism may lie in the aspect that LBP can increase the SAM content by reshaping the intestinal flora, resulting in DNA hypermethylation and suppressed expression of certain genes related to RA, thus ameliorating RA. Specifically, the LBP intervention reduced the abundances of Lachnospiraceae_NK4A136_group and uncultured_ bacterium_f_Rumino-coccaceae, and increased the abundances of Romboutsia, Lactobacillus, Dubosiella and Faecalibaculum. The content of the direct methyl donor SAM possibly generated from the above elevating gut bacteria was raised, and subsequently increased the methylation degree of specific colonic epithelial genes reportedly associated with RA including Dpep3, Gstm6, Slc27a2, Col11a2, Sycp2, SNORA22, Tnni1, Gpnmb, Mypn and Acsl6, and further inhibited their expressions. The symptoms of RA including the paw swell, joint pathological change and inflammatory status were eventually improved possibly through the gut-joint axis. The current study lacked the direct confirmation on the capability of SAM production by those bacteria with increased relative abundance, as well as the elucidation about the undefined function during RA alleviation of several genes. Therefore, further relevant studies are still needed in the future.
Methods
Reagents and materials
LBP was purchased from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). Our previous report have shown that the monosaccharide composition ratio of LBP is Mannose: Ribose: Rhamnose: Glucuronic acid: Galacturonic acid: Glucose: Galactose: Xylose: Arabinose=6:1:2:1:9:38:10:12:2146. Incomplete Freund’s adjuvant (IFA) and bovine type II collagen were purchased from Beijing Chondrex International Trade Co., Ltd. (Beijing, China).
Animal housing
All procedures were approved by the Institutional Animal Care Committee of Jinan University (2019827-02), and in accordance with the approved guidelines. Twenty-four specific-pathogen-free (SPF) female Wistar rats (8 weeks old) were purchased from Laboratory Animal Center of Southern Medical University (Guangzhou, Guangdong, China). Rats were raised in controlled standard barrier environment with temperature control (23 ± 2 °C), humidity control (55 ± 5%) and a 12-h light/12-h dark cycle at the Animal Center of Jinan University. During the experiment, rats were allowed to eat (AIN-93M) and drink (sterile distilled water) freely.
Induction of the type-II collagen arthritis models
According to the instructions of manufacturer, an equal volume of bovine type II collagen was mixed with IFA. The above mixture was totally emulsified by shaking before used. Rats were immunized with two subcutaneous injections with an interval of 7 days. For the first time, 0.2 mL of emulsion was injected at 1.5 cm away from the caudal end of the rats; and for the second time, 1.5 mL of the emulsion was injected subcutaneously at 3 cm from the caudal end of the rats.
Grouping, treatment and sample preparation
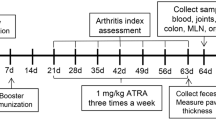
After 7 days of adaption, rats were randomly divided into three groups (n = 8 for each), which were the Control, Model and Lyc groups. For rats in the Lyc group, LBP solution (400 mg·kg−1) was given once a day by gavage, for those in the Control and Model groups, equal volume of sterilized distilled water was used to replace the LBP solution, and the administration of LBP lasted for 42 days (Fig. 7)52.
After adaptive feeding, bovine type II collagen and IFA mixture were subcutaneous injected to rats at day 7 for primary immunization, and at day 14 for booster immunization. The experiment lasted for 42 days, and the LBP solution and sterilized distilled water were orally administered to rats during this period.
The dose of 400 mg·kg−1 body weight for rats is equivalent to the dose of 63.49 mg·kg−1 body weight for human. On day 7 and 14, the rats were immunized by bovine type II collagen and IFA mixture according to the methods described above to establish RA models (the rats in the Control group were injected with the equivalent saline). On day 42, the rats were fasted overnight, and anesthetized with pentobarbital sodium on day 43 by intraperitoneal injection (50·mg kg−1 body weight) (Qiyun, Guangzhou, Guangdong, China). The blood samples were collected from the abdominal aorta and then the serum samples were obtained by centrifuged at 12,000 rpm for 30 min. After the rats were sacrificed, the ceca were removed and the cecal content of each sample was transferred into a sterile EP tube. Joint soft tissues were also removed and then packed with the saline-rinsed gauze. Colonic epithelial tissues were scraped from the inner side of colon, placed into the sterile folded silver paper and then stored in the liquid nitrogen immediately. All samples except the colonic epithelial tissues were stored in a cryogenic refrigerator at −80 °C.
Assessment of CIA
The diameter of rat’s hind paws, which is one of the indices to evaluate the severity of CIA model, were measured with a vernier caliper every 7 days. From the 7th day, the arthritis score which graded from 0 to 4 were recorded with the same frequency to the measurement of hind paw diameters53. The scoring criteria was listed at Table 3.
HE staining and pathological evaluation
After euthanasia, the rats’ hind feet were collected. The joint tissues were removed from the hind feet, fixed in 4% (v/v) neutral paraformaldehyde and then decalcified in 10% EDTA for 30 days. They were then embedded in paraffin. Joint tissues were sectioned at 5 μm of thickness, stained with HE. The HE-stained sections were evaluated in terms of inflammatory cell infiltration, synovial hyperplasia, fibrous tissue hyperplasia and pannus formation.
Measurement of cytokine contents in serum
The serum was stored at −80 °C until analysis. The contents of IL-1α, IL-1β, IL-10, IL-12 and IL-17 in serum were measured with ELISA kits according to the manufacturer’s instructions (Nanjing SenBeiJia Biological Technology Co., Ltd, Nanjing, China and Jiangsu Enzyme Industry Co., Ltd., China).
16S rDNA sequencing and bioinformatics analysis
Based on the previous research, the cecal bacterial DNA of each rat was extracted and analyzed54. Briefly, V3-V4 regions of the bacterial 16S rRNA gene were amplified by polymerase chain reaction (PCR), then PCR amplification products were purified, quantified and homogenized to construct sequencing libraries. The Illumina HiSeq Sequencing Platform (Illumina HiSeq 2500) was applied to sequence the library that pass the quality inspection. Then the library was converted into the original sequenced sequence (Sequenced Reads) by Base Calling analysis, splicing and filtering Reads, clustering OTUs, and performing species annotation and abundance analysis, alpha diversity analysis (Alpha Diversity), beta diversity analysis (Beta Diversity) and significant species difference analysis.
Analysis of transcriptome in colonic epithelium
Total RNA was extracted with RNeasy Mini Kit (QIAGEN, Shanghai, China), and RNA integrity (RIN) was evaluated with an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, USA). The qualified total RNA was purified with RNAClean XP Kit (Beckman Coulter, Shanghai, China) and RNase-Free DNase Set (QIAGEN, Shanghai, China). The mRNA was isolated from purified total RNA and the genomic mRNA was fragmented. The cDNA was synthesized with random hexamers and then subjected to end repair. The libraries were size-selected for cDNA target fragments of 200–300 bp with 2% low-range ultra-agarose gels followed by PCR amplification using Phusion DNA polymerase for 15 PCR cycles. After quantification using a Qubit® 2.0 Fluorometer and Agilent Bioanalyzer 2100, the paired-end RNA-seq sequencing library was sequenced with the IlluminaHiSeq 4000.
To identify the DEGs between two different groups, the expression level of each transcript was calculated according to the FRKM method. DEGs were selected with the following criteria: P value should be less than 0.05 and the absolute value of base-2 logarithm to fold change should be larger than 2. The R statistical package software Empirical Analysis of Digital Gene Expression in R (EdgeR) was utilized for the differential expression analysis.
Analysis of methylome in colonic epithelial cells
DNA from colonic epithelial cell samples was extracted with a gDNA Extraction Kit (Omega, Guangzhou, Guangdong, China). Then, the recommended Methylation-Sequencing protocol and specific enzymes were used to repair the ends, adenylate the 3’ ends and ligate methylated adaptors. The samples were then hybridized to biotinylated RNA baits and purified with streptavidin beads (New England BioLabs, Beijing, China). The remaining target sequences were bisulfite converted with an EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA, USA) as described in the Methylation-Sequencing protocol. The sequencing library was established by PCR amplification. Finally, the libraries were indexed and randomly pooled into multiplexes of four samples, and then sequenced using one pool per lane on the Illumina Hiseq X ten Platform (150 bp paired-end reads, Illumina Technologies, Shanghai, China). Sequencing was completed at the Shanghai Biotechnology Corporation (Shanghai, China).
The quality of raw data was evaluated with FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The sequencing quality (Q) was calculated with the formula of Q = −10Log10E, where E stands for the sequencing error rate. The clean reads were obtained after the filtration using Trim_galore v0.4.1 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with the following procedures: 1) remove the reads with low quality (Q < 20 in 3’ end or Q > 20 but accounted for less than 50%); 2) remove all adaptor sequences inside the reads; 3) remove reads with length less than 70 and unpaired. The clean reads of genome were compared by Bismark v0.15.0 using the genome of Rattus norvegicus as reference. 55. The methylation sites were scanned by Bison v0.4.0 with default setting56. Differential methylation sites (DMS) and DMR were detected and annotated by the tool of dispersion Shrinkage for Sequencing data (DSS) in the Bioconductor package57. Those sites with P < 0.05 and difference of methylation degree>10% were considered as DMS, and the region within which all CpG sites were with P < 0.05 was considered as DMR. The DMS were mapped into Gene Ontology (GO) database, and screened by hypergeometric test. Significant enrichment can be considered when P < 0.05.
Detection of S-adenosyl methionine (SAM) content in colonic epithelial tissue and intestinal contents
The mixture of 1 g colonic epithelial tissue and 200 μL saline or 0.1 g intestinal contents and 900 μL saline was fully homogenized with a high-throughput tissue homogenizer, and centrifuged at 3000 rpm for 15 min, and the supernatant was then carefully collected. The content of SAM was measured by corresponding ELISA kit according to the manufacturer’s instructions (Jiangsu Enzyme Industry Co., Ltd., China). The epithelial samples and the standards were added to the enzyme-labeled coating plate. The plate was incubated at 37 °C for 30 min, and then repeatedly washed with the washing solution for 5 times. After that, 50 μL of HRP-conjugate reagent was added to the plate. The plate was incubated at 37 °C for 30 min and washed for 5 more times. After that, 50 μL of chromogenic solution A and chromogenic solution B were added to the plate, and then the plate was kept in dark for ten minutes at 37 °C. Finally, the stop solution was added into the plate, and the absorbance was detected at the wavelength of 450 nm with a grating-type multifunctional microplate detector.
Statistical analysis
The statistical analysis of the bioinformatics data was performed using R software v3.3.1 with the default setting. Other analysis was performed with SPSS 20.0 software. The results were presented as the mean values with standard deviations. The normal distribution of data was checked by Shapiro-Wilk test when the significance was larger than 0.05. One-way analysis of variance (ANOVA) with Tukey’s test was applied to analyze the significance among different groups. Correlation heatmap was generated using the software of Origin 2021. Statistical significance was set at a P value less than 0.05.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA844171 and PRJNA846997. The other data from the current study are available from the corresponding authors on reasonable request.
References
Jiang, J. B. et al. Therapeutic effects of astragalus polysaccharides on inflammation and synovial apoptosis in rats with adjuvant-induced arthritis. Int. J. Rheum. Dis. 13, 396–405 (2010).
Wang, Q. et al. Curcumin attenuates collagen-induced rat arthritis via anti-inflammatory and apoptotic effects. Int. Immunopharmacol. 72, 292–300 (2019).
Xiao, J., Li, S., Wang, W., Li, Y. & Zhao, W. Protective effects of overexpression TCR V beta 5.2-HSP70 and TCR V beta 8.2-HSP70 against collagen-induced arthritis in rats. Cell. Mol. Immunol. 4, 439–445 (2007).
Liu, L., Sha, X. Y., Wu, Y. N., Chen, M. T. & Zhong, J. X. Lycium barbarum polysaccharides protects retinal ganglion cells against oxidative stress injury. Neural Regen. Res. 15, 1526–1531 (2020).
Tian, B. et al. Dietary whole Goji berry (Lycium barbarum) intake improves colonic barrier function by altering gut microbiota composition in mice. Int. J. Food Sci. Tech. 56, 103–114 (2020).
Ran, L. et al. Antitumor effects of pollen polysaccharides from Chinese wolfberry on DU145 cells via the PI3K/AKT pathway in vitro and in vivo. Int. J. Biol. Macromol. 152, 1164–1173 (2020).
Li, W., Gao, M. & Han, T. Lycium barbarum polysaccharides ameliorate intestinal barrier dysfunction and inflammation through the MLCK-MLC signaling pathway in Caco-2 cells. Food Funct. 11, 3741–3748 (2020).
Fu, Y. W. et al. Lycium barbarum polysaccharide-glycoprotein preventative treatment ameliorates aversive. Neural Regen. Res. 16, 543–549 (2021).
Liu, Y. et al. Lycium barbarum polysaccharide attenuates type II collagen-induced arthritis in mice. Int. J. Biol. Macromol. 78, 318–323 (2015).
Cai, S., Sun, J. & Wei, X. Lycium barbarum polysaccharide inhibits NF-kappaB pathway to reduce the level of inflammatory cytokines in osteoarthritis chondrocytes (in Chinese). Chin. J. Cell. Mol. Immunol. 34, 989–993 (2018).
Kalinkovich, A. & Livshits, G. A cross talk between dysbiosis and gut-associated immune system governs the development of inflammatory arthropathies. Semin. Arthritis Rheu. 49, 474–484 (2019).
Dourado, E., Ferro, M., Sousa Guerreiro, C. & Fonseca, J. E. Diet as a modulator of intestinal microbiota in rheumatoid arthritis. Nutrients 12, 3504 (2020).
Van de Wiele, T., Van Praet, J. T., Marzorati, M., Drennan, M. B. & Elewaut, D. How the microbiota shapes rheumatic diseases. Nat. Rev. Rheumatol. 12, 398–411 (2016).
Zhuo, C. et al. Schizophrenia and gut-flora related epigenetic factors. Prog. Neuro-Psychoph. 90, 49–54 (2019).
Liu, J., Liu, J., Liu, L., Zhang, G. & Peng, X. Reprogrammed intestinal functions in Astragalus polysaccharide-alleviated osteoporosis: combined analysis of transcriptomics and DNA methylomics demonstrates the significance of the gut-bone axis in treating osteoporosis. Food Funct. 12, 4458–4470 (2021).
Ye, L. et al. IL-37 alleviates rheumatoid arthritis by suppressing IL-17 and IL-17-triggering cytokine production and limiting Th17 cell proliferation. J. Immunol. 194, 5110–5119 (2015).
Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Bio. 20, 590–607 (2019).
Moore, L. D., Le, T. & Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38 (2013).
Gilbert, J. A. et al. Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018).
Miro-Blanch, J. & Yanes, O. Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front. Genet. 10, 638 (2019).
Taneja, V. Cytokines pre-determined by genetic factors are involved in pathogenesis of Rheumatoid arthritis. Cytokine 75, 216–221 (2015).
Zhang, X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015).
Horta-Baas, G. et al. Intestinal dysbiosis and rheumatoid arthritis: a link between gut microbiota and the pathogenesis of rheumatoid arthritis. J. Immunol. Res. 2017, 4835189 (2017).
Goh, C. E., Kopp, J., Papapanou, P. N., Molitor, J. A. & Demmer, R. T. Association between serum antibodies to periodontal bacteria and rheumatoid factor in the third national health and nutrition examination survey. Arthritis Rheumatol. 68, 2384–2393 (2016).
Rajilic-Stojanovic, M. & de Vos, W. M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 38, 996–1047 (2014).
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013).
Vacca, M. et al. The controversial role of human gut. Lachnospiraceae. Microorg. 8, 573 (2020).
Kronzer, V. L., Crowson, C. S., Sparks, J. A., Myasoedova, E. & Davis, J. M. III Comorbidities as risk factors for rheumatoid arthritis and their accrual after diagnosis. Mayo Clin. Proc. 94, 2488–2498 (2019).
Wu, X. et al. An integrated microbiome and metabolomic analysis identifies immunoenhancing features of Ganoderma lucidum spores oil in mice. Pharmacol. Res. 158, 104937 (2020).
Pan, H. et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome 7, 107 (2019).
Liu, X. et al. Lactobacillus salivarius isolated from patients with rheumatoid arthritis suppresses collagen-induced arthritis and increases Treg frequency in mice. J. Interf. Cytok. Res. 36, 706–712 (2016).
Esvaran, M. & Conway, P. L. Lactobacillus fermentum PC1 has the capacity to attenuate joint inflammation in collagen-induced arthritis in DBA/1 mice. Nutrients 11, 785 (2019).
Yamashita, M. et al. Preventive effect of Lactobacillus helveticus SBT2171 on collagen-induced arthritis in mice. Front. Microbiol. 8, 1159 (2017).
Nasir, A. et al. Nutrigenomics: Epigenetics and cancer prevention: A comprehensive review. Crit. Rev. Food Sci. Nutr. 60, 1375–1387 (2020).
Huang, W. Y. et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res 43, D856–D861 (2015).
Lawrence, E. A. et al. The mechanical impact of col11a2 loss on joints; col11a2 mutant zebrafish show changes to joint development and function, which leads to early-onset osteoarthritis. Philos. Trans. R. Soc. B Biol. Sci. 373, 20170335 (2018).
Rice, S. J., Cheung, K., Reynard, L. N. & Loughlin, J. Discovery and analysis of methylation quantitative trait loci (mQTLs) mapping to novel osteoarthritis genetic risk signals. Osteoarthr. Cartil. 27, 1545–1556 (2019).
Teodoro, B. G. et al. Long-chain acyl-CoA synthetase 6 regulates lipid synthesis and mitochondrial oxidative capacity in human and rat skeletal muscle. J. Physiol.-Lond. 595, 677–693 (2017).
MacDonald, I. J. et al. Implications of angiogenesis involvement in arthritis. Int. J. Mol. Sci. 19, 2012 (2018).
Karlsson, C. et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthr. Cartil. 18, 581–592 (2010).
Muragaki, Y. et al. Alpha 1(VIII)-collagen gene transcripts encode a short-chain collagen polypeptide and are expressed by various epithelial, endothelial and mesenchymal cells in newborn mouse tissues. Eur. J. Biochem. 207, 895–902 (1992).
Rose, A. A. N. et al. ADAM10 releases a soluble form of the GPNMB/Osteoactivin extracellular domain with angiogenic properties. PLoS One 5, e12093 (2010).
Taniuchi, K. & Ogasawara, M. KHSRP-bound small nucleolar RNAs associate with promotion of cell invasiveness and metastasis of pancreatic cancer. Oncotarget 11, 131–147 (2020).
Caimari, A., Oliver, P., Rodenburg, W., Keijer, J. & Palou, A. Slc27a2 expression in peripheral blood mononuclear cells as a molecular marker for overweight development. Int. J. Obes. 34, 831–839 (2010).
Cifre, M. et al. Human peripheral blood mononuclear cell in vitro system to test the efficacy of food bioactive compounds: Effects of polyunsaturated fatty acids and their relation with BMI. Mol. Nutr. Food Res. 61, 1600353 (2017).
Saade, M., de Souza, G. A., Scavone, C. & Kinoshita, P. F. The role of GPNMB in inflammation. Front. Immunol. 12, 674739 (2021).
Zhang, Q. et al. Landscape and dynamics of single immune cells in hepatcellular carcinoma. Cell 179, 829–845 (2019).
Sato, W. et al. Hepatic gene expression in hepatocyte-specific Pten deficient mice showing steatohepatitis without ethanol challenge. Hepatol. Res. 34, 256–265 (2006).
Hu, B., Yang, X. B. & Sang, X. T. Construction of a lipid metabolism-related and immune-associated prognostic signature for hepatocellular carcinoma. Cancer Med. 9, 7646–7662 (2020).
Tillmann, S. et al. Probiotics affect one-carbon metabolites and catecholamines in a genetic rat model of depression. Mol. Nutr. Food Res. 62, e1701070 (2018).
Chu, J., Qian, J., Zhuang, Y., Zhang, S. & Li, Y. Progress in the research of S-adenosyl-L-methionine production. Appl. Microbiol. Biot. 97, 41–49 (2013).
Wang, C. et al. Recovery of Ggt7 and Ace expressions in the colon alleviates collagen-induced arthritis in rats by specific bioactive polysaccharide intervention. J. Agric. Food Chem. 68, 14531–14539 (2020).
Fan, Z. et al. The prophylactic effects of different Lactobacilli on collagen-induced arthritis in rats. Food Funct. 11, 3681–3694 (2020).
Zheng, C. et al. Impact and consequences of polyphenols and fructooligosaccharide interplay on gut microbiota in rats. Food Funct. 8, 1925–1932 (2017).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Ryan, D. P. & Ehninger, D. Bison: bisulfite alignment on nodes of a cluster. BMC Bioinforma. 15, 337 (2014).
Park, Y. & Wu, H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 32, 1446–1453 (2016).
Acknowledgements
This work was supported by the National Natural Science Funds (No. 31801543 and No. 32072223), the Natural Science Funds of Guangdong Province, China (No. 2022A1515011344), and the Funding by Science and Technology Projects in Guangzhou (202201020072).
Author information
Authors and Affiliations
Contributions
X.P. designed and guided the project; W.L. and C.W. performed the animal experiment; C.W., and J.L. collected the samples; J.L. and C.W. performed the data analysis; C.W., W.L., R.L. and J.L. wrote the paper; X.P. and J.L. revised the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lai, W., Wang, C., Lai, R. et al. Lycium barbarum polysaccharide modulates gut microbiota to alleviate rheumatoid arthritis in a rat model. npj Sci Food 6, 34 (2022). https://doi.org/10.1038/s41538-022-00149-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41538-022-00149-z