Abstract
After traumatic injury, healing of mammalian ligaments is typically associated with fibrotic scarring as opposed to scar-free regeneration. In contrast, here we show that the ligament supporting the jaw joint of adult zebrafish is capable of rapid and complete scar-free healing. Following surgical transection of the jaw joint ligament, we observe breakdown of ligament tissue adjacent to the cut sites, expansion of mesenchymal tissue within the wound site, and then remodeling of extracellular matrix (ECM) to a normal ligament morphology. Lineage tracing of mature ligamentocytes following transection shows that they dedifferentiate, undergo cell cycle re-entry, and contribute to the regenerated ligament. Single-cell RNA sequencing of the regenerating ligament reveals dynamic expression of ECM genes in neural-crest-derived mesenchymal cells, as well as diverse immune cells expressing the endopeptidase-encoding gene legumain. Analysis of legumain mutant zebrafish shows a requirement for early ECM remodeling and efficient ligament regeneration. Our study establishes a new model of adult scar-free ligament regeneration and highlights roles of immune-mesenchyme cross-talk in ECM remodeling that initiates regeneration.
Similar content being viewed by others
Introduction
Ligament injuries typically heal with biomechanically inferior scar tissue that compromises joint stability and increases the risk of developing painful and debilitating osteoarthritis1,2. Healed ligaments fail to recapitulate the highly organized structure of the uninjured tissue. Foundational research using non-regenerative models has highlighted that the failure to regenerate ligaments without scarring is common among mammals3,4,5. Early studies investigating repair of the medial collateral ligament (MCL) following rupture showed that the healed ligament undergoes more deformation under the same load, coincident with disorganization of the collagen fibers at the wound site5,6. The first 10 days of ligament healing in mammals are characterized by an invasion of fibroblasts, vasculature, and pro-inflammatory immune cells along the ligament6,7. While ligament repair continues for 14 weeks, an incomplete remodeling of both the cellular and extracellular matrix (ECM) composition occurs, ending with a fibrotic scar bridging the injury. Similarly, in human and murine tendon and ligament injuries, the stubs are bridged by α-Smooth Muscle Actin-positive scarring myofibroblasts8,9,10,11. Notably, these myofibroblasts do not express tendon and ligament marker Scleraxis (Scx), indicating that they are not true ligamentocytes8. Existing studies on adult connective tissue injury highlight the importance of the reestablishment of cell fate and ligament ECM through regeneration.
Recent advances in tendon injury modeling showed that neonatal mice are capable of scar-free regeneration after Achilles tendon transection8. In this neonatal repair context, Scx-positive tenocytes retain their developmental potency to proliferate, migrate to the wound site, and heal without forming a fibrotic scar8. Further studies on developmental regenerative potential have been performed using juvenile zebrafish, which described that BMP-dependent activation of perichondrial progenitors is necessary for tendon regeneration after tendon cell ablation injury12. However, while these models interrogate scar-free tendon regeneration in the context of development, there are currently no models to study scar-free ligament regeneration in mature adult ligaments. To address this need, we utilized the highly regenerative zebrafish, as they are a well-established model for heart, spinal cord, and tail fin regeneration13,14,15 and moreover possess regenerative synovial joints supported by ligaments16,17,18.
In the course of establishing a post-traumatic osteoarthritis model, we had observed rapid regeneration of the craniofacial interopercular-mandibular (IOM) ligament, which supports the zebrafish jaw joint, following complete transection in one-year-old fish18. Here we characterize the cellular and molecular events underlying the three overlapping phases of ligament healing: inflammation, proliferation, and remodeling. We also used single-cell RNA sequencing (scRNAseq) to characterize the dynamics of cell populations during regeneration and identified legumain (lgmn) as a gene highly enriched in macrophages that invade the injury site. Legumain is a member of the C13 family of cysteine proteases that regulate ECM factors such as cathepsins, fibronectin1 (Fn1), and matrix metalloprotease-2 (MMP-2)19,20. Through injury analyses in a zebrafish mutant for lgmn, we uncovered an essential role in regulating ECM remodeling and scar-free ligament regeneration. Our findings highlight the important role of immune-mediated ECM remodeling for successful scar-free regeneration.
Results
Complete and scar-free regeneration of the IOM ligament following transection
The IOM ligament connects the interopercular bone to the retroarticular bone of the mandible to control movement of the zebrafish jaw joint. Following complete transection of the IOM ligament in adult zebrafish, a time-course of histological analysis revealed dynamic morphological changes during ligament regeneration (Fig. 1a–h). The uninjured ligament contains ligamentocytes within a cell-sparse ECM (Fig. 1a, b). Within 1 day post-ligament transection (dplt), a robust inflammatory response is seen that begins to resolve by 3 dplt. From 1 to 3 dplt, we observe breakdown of ligament ECM and rounding of ligament-embedded nuclei (Fig. 1c, d). Beginning at 3 dplt and continuing through 7 dplt, abundant mesenchymal cells are observed in the gap between the injured ligament stubs (Fig. 1d, e). By 14 dplt, cells bridging the RA and IOP bones display an elongated morphology in the direction of force but at a high cell density compared to native uninjured ligaments (Fig. 1f). From 21 to 28 dplt, the hypercellularity is resolved and the regenerated ligament resembles the cell-sparse uninjured ligament (Fig. 1g, h). These data point to a transient inflammatory response and ligament stub ECM breakdown, followed by establishment of a population of mesenchymal cells bridging the wound site that are remodeled into the regenerated ligament.
a Schematic of the articulating bones at the zebrafish jaw joint and connections of the interopercular-mandibular (IOM) ligament. b–h Toluidine blue staining of adult zebrafish IOM ligament in uninjured (b, n = 16), 1 day post-ligament transection (dplt, c, n = 3), 3 dplt (d, n = 5), 7 dplt (e, n = 6), 14 dplt (f, n = 16), 21 dplt (g, n = 3), and 28 dplt (h, n = 16). i–n Repeated live imaging of the ventral view of scxa:mCherry expression in IOM ligament before injury (i, n = 9), immediately after injury (j, n = 3), 3 dplt (k, n = 9), 6 dplt (l, n = 3), 14 dplt (m, n = 9), and 28 dplt (n, n = 3). o–r Confocal microscopy imaging of tissue cleared thbs4a_p1:GFP expression in IOM ligament uninjured control (o, n = 6), 1 dplt (p, n = 5), 7 dplt (q, n = 6), and 28 dplt (r, n = 3). AA, anguloarticular bone; IOM, interopercular-mandibular ligament; IOP, interopercular bone; Q, quadrate bone; RA, retroarticular bone. Scale bars = 100 μm (b–h, o–r), 250 μm (i–n).
To understand the activity of ligamentocytes during regeneration, we labeled these cells with a scxa:mCherry transgenic line. In these fish, mCherry expression is driven by the regulatory region surrounding the scleraxis a (scxa) gene, which encodes a transcription factor highly expressed in ligament and tendon cells21,22. In uninjured adults, live imaging revealed scxa:mCherry fluorescence in each of the bilateral IOM ligaments (Fig. 1i). Immediately following the transection, we observed decreased scxa:mCherry fluorescence only on the injured side, with mCherry fluorescence nearly completely absent by 3 dplt (Fig. 1j, k). We then saw reappearance of scxa:mCherry fluorescence by 6 and 14 dplt, gradually returning to near-normal levels by 28 dplt (Fig. 1l–n). This result suggests that while ligament stub-resident ligamentocytes were observed in histological analysis, these mature cells show a dramatic loss of scxa:mCherry expression adjacent to the transection site.
Expression of scxa is known to be broader than just tenocytes and ligamentocytes, including early mesenchyme and fibroblasts23. We therefore sought to confirm our results with a transgenic line marking mature ligamentocytes more specifically. Analysis of our published single-cell assay for transposase-accessible chromatin (scATAC) sequencing data of zebrafish cranial neural crest-derived cells24 identified a 635 bp region within intron 13 of the thrombospondin 4a (thbs4a) gene that was selectively accessible in the tendon and ligament cluster (Supplementary Fig. 1a). We then used this sequence to generate thbs4a_p1:eGFP zebrafish. In contrast to expression of scxa:mCherry throughout tendons and ligaments, thbs4a_p1:eGFP expression was highly specific for the IOM ligament, with the exception of a small amount of superficial cartilage expression in the jaw joint (Fig. 1o; Supplementary Fig. 1b). Following IOM ligament transection, we observed disappearance of thbs4a_p1:eGFP expression in spared ligament domains flanking the cut site by 1 dplt, reappearance by 7 dplt, and remodeling to a normal ligament morphology by 28 dplt (Fig. 1o–r, Supplementary Videos 1-4). In particular, the use of CUBIC tissue clearing technique revealed that ligamentocytes had re-established alignment in the direction of force at 28 dplt, in contrast to the disorganized alignment seen at 7 dplt. The loss of expression of two independent reporters in ligamentocytes flanking the transection site strongly suggest that remaining ligamentocytes dedifferentiate early in the regenerative process. Further, re-expression of these reporters later in the regenerative process show re-activation of the developmental gene regulatory program for ligamentocytes during adult regeneration.
Mature ligamentocytes contribute to the regenerated ligament
Next, we sought to define the source of ligamentocytes during regeneration. During development, the IOM ligament and other components of the craniofacial skeleton are neural crest-derived. We therefore tested whether the regenerative mesenchyme and regenerated ligament are similarly neural crest-derived. To do so, we permanently labeled neural crest-lineage cells in Sox10:Cre;actb2:loxP-BFP-STOP-loxP-DsRed fish, in which Sox10-driven Cre recombinase switches blue fluorescent protein (BFP) to DsRed fluorescent protein (Fig. 2a). Repeated live imaging through the course of ligament healing revealed the regenerative mesenchyme to be DsRed+ at 3, 7, and 14 dplt, and the regenerated ligament DsRed+ at 14 and 28 dplt. Immunofluorescence staining of tissue sections against DsRed protein highlight DsRed expression in the uninjured ligament and skeletal mesenchyme, as well as conversion of the bridging regenerative mesenchyme and injured ligamentocytes at 3 dplt (Fig. 2b,c). Thus, similar to their development, both the early bridging regenerative mesenchyme and regenerated ligament are produced by neural crest-derived cells.
a Lineage trace of DsRed-labeled cells using Sox10:Cre with actb2:loxP-BFP-STOP-loxP-DsRed. (b, c) DsRed immunofluorescence (magenta) in Sox10:Cre;actb2:loxP-BFP-STOP-loxP-DsRed from uninjured (b) and 3 dplt (c) (n = 4/time-point). Insets highlight NC-derived cells in periosteum (b’), uninjured ligament (b”), regenerative mesenchyme (c’), and injured ligament (c”). Ligament outlined in white dashed line. d Schematic of ligamentocyte lineage tracing. thbs4a_p1:CreER;actb2:loxP-BFP-STOP-loxP-DsRed fish were 4-OHT treated, screened for conversion, then imaged. e Repeated ventral live imaging of DsRed-labeled thbs4a-lineage before, immediately following, and 28 dplt (cut site at dashed white line) (n = 5/time point). DsRed (magenta) and thbs4a (cyan) smFISH in 28 dplt (f), uninjured (g) and 3 dplt (h) ligaments from thbs4a_p1:CreER;actb2:loxP-BFP-STOP-loxP-DsRed (n = 5/time point). Insets illustrate DsRed single-positive (h’), thbs4a single-positive (h”), and DsRed-thbs4a double-positive cells in the regenerative mesenchyme (h”’). DsRed (magenta), col12a1a (yellow), and fn1a (cyan) smFISH in uninjured (i) and 3 dplt (j) converted thbs4a_p1:CreER;actb2:loxP-BFP-STOP-loxP-DsRed fish (n = 5). Insets illustrate DsRed single-positive (j’) and fn1a-col12a1a double-positive regenerative mesenchyme (j”), and DsRed-fn1a double-positive cells in the ligament (j”’). PCNA immunofluorescence (green) in uninjured (k), 1 dplt (l), and 3 dplt (m). Ligament outlined in white dashed line, regenerative mesenchyme domain in red dashed line. n Quantification of percent PCNA+ nuclei in (k–m), showing significant increase in proliferation at 3 dplt (n = 4-6 per time point). Scale bars = 500 μm (a, e), 50 μm (b, c, g, h, I, j), 20 μm (b’, c’, h’, j’).
We next sought to understand which subset of neural crest-derived cells might contribute to the regenerated ligament. Given our finding that mature ligamentocytes undergo rapid dedifferentiation in response to ligament transection, we tested their potential long-term contribution to the regenerated ligament. To permanently label ligamentocytes prior to injury, we generated a transgenic line that used the thbs4a_p1 ligament-specific enhancer to drive expression of 4-hydroxy tamoxifen (4-OHT)-inducible CreERT2 (thbs4a_p1:CreERT2) in combination with the ubiquitously-expressed actb2:loxP-BFP-STOP-loxP-DsRed reporter. Treatment of adult double transgenic fish with 3 doses of 4-OHT resulted in DsRed fluorescence throughout the IOM ligaments (Fig. 2d, e). Following ligament transection, repeated live imaging revealed the regenerated ligament was DsRed+ at 28 dplt, with DsRed expression co-localizing with endogenous expression of thbs4a in ligamentocytes (Fig. 2e, f).
To further investigate the dynamic fate of the ligamentocyte lineage during regeneration, we assessed the co-localization of DsRed with mature ligamentocyte marker thbs4a. In situ hybridization highlights that while DsRed transcripts colocalize with thbs4a transcripts in the uninjured adult ligament, DsRed-positive cells appear in the regenerative mesenchyme at 3 dplt that do not express thbs4a (Fig. 2g, h). thbs4a single-positive cells are also present, suggesting the initiation of a ligamentocyte lineage program from either unconverted ligamentocytes or non-ligament progenitors. Consistent with live imaging of scxa:mCherry and thbs4a_p1:eGFP, these data show that pre-existing ligamentocytes de-differentiate, repressing markers of mature ligamentocyte identity as they migrate into the wound.
To characterize the identity of the ligamentocyte-derived regenerative mesenchyme, we assessed the expression of markers fibronectin 1a (fn1a) and collagen 12 alpha 1a (col12a1a), which label injury mesenchyme in zebrafish spinal cord and heart regeneration25,26,27,28. While fn1a is minimally expressed in the uninjured ligament, we see abundant fn1a transcripts in DsRed+ thbs4a-lineage cells in both the injured ligament stubs, as well as in the bridging regenerative mesenchyme (Fig. 2i, j). Additionally, we see high expression of col12a1a transcripts in thbs4a-lineage cells in the regenerative mesenchyme relative to the uninjured ligament. Together, these data indicate that thbs4a-lineage cells lose characteristic markers of ligament fate and undergo dynamic changes in their ECM gene expression profile as they migrate into the wound and bridge the ligament stubs.
In addition, we observed extensive proliferation, based on immunofluorescence staining for Proliferating Cell Nuclear Antigen (PCNA), within both the regenerative mesenchyme and ligament stubs adjacent to the transection site. Compared to only minimal PCNA+ cells prior to injury and at 1 dplt, 39% of cells within the ligament stub domain and 47.5% cells within the regenerative mesenchyme domain were PCNA+ at 3 dplt (Fig. 2k–n, n = 4-6 per sample). These data support typically quiescent ligamentocytes re-entering the cell cycle upon injury and contributing to the regenerated ligament.
Single-cell RNA sequencing reveals dynamic mesenchymal populations during ligament regeneration
To gain insights into the cellular and molecular dynamics underlying ligament regeneration, we performed droplet-based single-cell RNA sequencing (scRNAseq) on micro-dissected IOM ligaments, including adjacent joint and mesenchymal tissues, before and during early stages of regeneration. We used FACS in Sox10:Cre;actb2:loxP-BFP-STOP-loxP-DsRed fish to enrich for neural crest-derived cells, yet sequenced both DsRed+ neural crest-derived and BFP+ non-neural crest-derived populations to capture the entire diversity of cell types during regeneration. DsRed+ and BFP+ populations were sequenced independently from uninjured controls, and from fish at 1 and 3 dplt or undergoing sham surgery to cut the skin but not the ligament (Fig. 3a). After pre-processing and filtering, we obtained 9,451 cells (1618 DsRed + , 7833 BFP + ) with a median 766 genes per cell (Supplementary Table 1).
a Schematic detailing the generation and analysis of single-cell RNA sequencing data. Jaw joints from uninjured, 1 day post-SHAM (nicking of the epithelium above the ligament) surgery, 3 days post-SHAM surgery, 1 day post-ligament transection, and 3 days post-ligament transection were dissociated and sorted for DsRed (neural crest-derived) or BFP (non-neural crest-derived). b UMAP of low-resolution superclusters for merged samples. Superclusters include 2 neural crest-derived clusters (NC-Derived), 2 immune clusters, 3 epithelial clusters, and 1 endothelial cluster. c UMAP colored for neural crest-lineage enrichment. Cells from FACS-sorted DsRed+ neural crest-derived cells colored in red, cells from sorted BFP+ non-neural crest-derived cells colored in blue. d DotPlot detailing expression of marker genes for each supercluster.
Unbiased, low-resolution clustering categorized cells into eight broad superclusters representing immune, epithelial, neural crest-derived (NC-derived), and endothelial cells (Fig. 3b, c). Epithelial superclusters included basal and intermediate keratinocytes (Epithelium 1, krt5+), superficial cells (Epithelium 2, krt4+), and mucosal cells (Epithelium 3, muc5.1+) (Supplementary Fig. 2). Immune 1 supercluster includes lymphoid and myeloid cells, such as T cells (cd8a+, lck+) and macrophages (mpeg1.1+, mfap4+), and Immune 2 cluster consisted primarily of neutrophils (mpx+, lyz+). We also detected an endothelial cluster (pecam1+, kdrl+). Two superclusters, enriched for DsRed+ cells (Fig. 3c), represent neural crest-derived mesenchymal populations marked by either the broad mesenchymal gene prrx1b (NC-Derived 1) or the chondrocyte genes col2a1a and mia (NC-Derived 2) (Fig. 3d).
Re-clustering of the two neural crest-derived superclusters revealed clusters of osteoblasts (ifitm5+), chondrocytes (col2a1a+), superficial chondrocytes and synoviocytes (prg4b+), ligamentocytes (scxa+, thbs4a+), two types of dermal fibroblasts (pah+), periosteal cells (mmel1+), three types of stromal cells (defined by either cxcl12a, coch, or col5a3a), and glial cells (mbpa+)18,24 (Fig. 4a–c, Supplementary Fig. 2, Supplementary Table 2). In addition, we identified two injury-specific clusters with stage-specific cell composition: 1 dplt regenerative mesenchyme (66% 1 dplt cells) and 3 dplt regenerative mesenchyme (82% 3 dplt cells). These regenerative mesenchyme clusters were characterized by high expression of collagen 12 alpha 1a (col12a1a) and fibronectin 1a (fn1a) (Fig. 4c; Supplementary Fig. 2). In situ hybridization shows that col12a1a is highly expressed in the mesenchymal cells bridging the ligament stubs by 3 dplt (Fig. 4d). Further, combinatorial in situ hybridizations show that a subset of col12a1a+ regenerative mesenchyme cells co-express the early ligamentocyte marker scxa, suggesting early initiation of the ligament program by 3 dplt (Fig. 4d). In contrast, col12a1a+ cells do not co-express alpha-smooth muscle actin 2 (acta2), a marker of pro-scarring fibroblasts in mammals. These data are consistent with regenerative mesenchyme cells producing new ligamentocytes as opposed to scar tissue during regeneration.
a, b UMAP of single-cell RNA sequencing data of neural crest-derived cells from early ligament regeneration colored by (a) cluster or (b) sample. c DotPlot of marker genes used to identify each cluster. d RNAscope single-molecule fluorescent in situ hybridization (smFISH) for scxa (cyan), col12a1a (yellow), and acta2 (magenta) in an uninjured ligament. d’ smFISH for scxa, col12a1a, and acta2 expression at 3 dplt. Insets highlighting scxa+ col12a1a+ regenerative mesenchyme (inset 1, n = 5 per time point), acta2+ perivascular cells (inset 2), and acta2-low mesenchyme near the ligament stub (inset 3). Scale bars = 50 μm (d, d’), 10 μm (insets in d), ligament stub outlined in white dashed line.
Single-cell profiling of the immune response to ligament injury
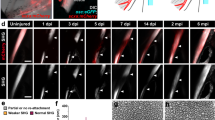
Re-clustering of the two immune cell superclusters revealed 11 clusters: two types of neutrophils (mpx+), macrophages (mpeg1.1+/mfap4+), dendritic cells (mpeg1.1+/mfap4-), two types of T helper cells (il21r.2+ or il4+), proliferative cells (mki67+), and four types of lymphocytes (lck+), including two T cell-containing clusters (cd8a+), NK cells (s1pr5a+)29, and activated NK cells (ifng1+/gzmk+) (Fig. 5a–c, Supplementary Fig. 3). The Neutrophil 1 cluster consisted predominantly of cells from the 1 dplt sample (82% of cells), indicative of an acute inflammatory phase shortly after injury. Repeated live imaging of individual fish expressing the transgenic neutrophil reporter mpx:mCherry revealed homing of mpx+ neutrophils to the injury site within 4 hours, with neutrophil accumulation beginning to resolve by 1 dplt (Fig. 5d, white arrowheads). In contrast, repeated imaging of fish transgenic for the macrophage and dendritic cell reporter mpeg1.1:eGFP showed mpeg1.1+ immune cells homing to the transection site at 1 dplt and resolving by 7 dplt (Fig. 5e, white arrowheads).
UMAPs of single-cell RNA sequencing datasets colored by cluster (a) and injury sample (b). c Dot Plot showing marker gene expression for each immune cluster. Repeated live imaging of neutrophils (d; mpx:mCherry) (n = 5) and macrophages (e; mpeg1.1:eGFP+) (n = 6) at the injury site (arrowheads) during the first week of regeneration color-coded by signal intensity. f Dot Plot demonstrating fn1a expression in macrophage cluster cells split by Sample and Feature Plot for 1 dplt-macrophage enriched gene fn1a. g Representative images of smFISH staining using fn1a (green) and mpeg1.1 (magenta) probes in IOM ligaments from uninjured and 1 dplt samples (n = 3). h Feature Plots for macrophage marker mpeg1.1 and macrophage-enriched gene lgmn. i smFISH for lgmn (yellow) and mpeg1.1 (magenta) probes in IOM ligaments from uninjured and 1 dplt samples (n = 3). Gray dashed lines outline the IOM ligament and inset positions are denoted by the numbers 1-2 (g, i). Scale bars = 500 μm (d, e), 50 μm (g, i).
Within the mpeg1.1+/mfap4+ macrophage cluster, we noted a subpopulation of macrophages with enriched composition of 1 dplt cells and selective expression of fn1a (Fig. 5f). In situ hybridization at 1 dplt revealed mpeg1.1+/fn1a+ macrophages close to the transection site; adjacent mpeg1.1-/fn1a+ cells likely represent regenerative mesenchyme cells that also express fn1a (Fig. 5g). In the uninjured IOM ligament, we observed almost no mpeg1.1+ cells or fn1a expression. One of the most selective markers of mpeg1.1+ cells was lgmn, and in situ hybridization revealed abundant mpeg1.1+/lgmn+ cells at 1 dplt but not in the uninjured IOM ligament (Fig. 5c, h, i; Supplementary Table 3). These data show that ligament transection induces a rapid recruitment of neutrophils that is followed by recruitment of lgmn+/fn1a+ macrophages to the injury site.
Legumain is required for efficient ligament regeneration
Given the high expression of lgmn in mpeg1.1+ immune cells that home to the ligament injury site, we tested the requirement for lgmn in ligament regeneration by using a premature stop allele (lgmnsa22350) (Fig. 6a). Homozygous lgmnsa22350 zebrafish (lgmn mutants) are anatomically similar to wild types with no overt differences in the gross morphology of their uninjured ligaments (Fig. 6b). To characterize ECM changes during ligament regeneration, we performed Acid Fuchsin Orange G (AFOG) staining in which bone and ligament ECM stains red, and cartilage and mesenchymal ECM stains blue (Fig. 6b). In wild type, we observed the disappearance of the normal red ligament ECM by 3 dplt, which then began to reappear by 7 and 14 dplt and was fully restored by 42 dplt. In lgmn mutants, ligament tissue adjacent to the transection site retained ligament ECM and was not resorbed at 3 dplt as in wild type (black arrow, Fig. 6b). By 14 dplt, we observed a striking delay in ligament regeneration compared to wild types, with lgmn mutants displaying disorganized non-ligament ECM throughout the injury domain. At 42 dplt, lgmn mutants displayed reduced and highly dysmorphic regenerated ligaments consisting of several distinct nodules that failed to align along the direction of force.
a Schematic of lgmn mutant locus showing a point mutation in exon 2 that encodes for an early stop codon. b Acid Fuchsin Orange-G (AFOG) histology staining of ECM remodeling in wild type and lgmn mutant IOM ligament regeneration in uninjured, 3 dplt, 7 dplt, 14 dplt, and 42 dplt. Black arrow denotes interface of injured ligament bundle with invading cells. Collagen is stained blue, protein deposits are stained red/purple, and cellular cytoplasm is stained orange (n = 3-5 per time point). c Representative images of PCNA immunofluorescence in wild type and lgmn mutants in uninjured and 3 dplt IOM ligaments. Region of interest for quantification is outlined in white dashed lines for the ligament and red dashed lines for regenerative mesenchyme domain. d Quantification of percentage of PCNA positive cells normalized with total DAPI-stained nuclei (n = 3-6 per time point); error bar represents SEM, p = 0.0161. Scale bars = 150 μm (b), 50 μm (c).
To examine the cellular bases of aberrant ligament regeneration in lgmn mutants, we first measured apoptosis and proliferation using TUNEL and PCNA immunofluorescence assays, respectively. After transection in wild types, we observed 7.4% TUNEL+ ligamentocyte nuclei at 1 dplt and 2.5% TUNEL+ at 3 dplt (Supplementary Fig. 4). Similar rates of apoptosis were seen in both wild types and lgmn mutants, suggesting that altered apoptotic load is not the initiator of fibrotic response in lgmn mutants (Supplementary Fig. 4). However, at 3 dplt, we observed a 27% decrease in proliferation within the regenerative mesenchyme domain of lgmn mutants (p = 0.0161) and a trend toward lower proliferation in ligament tissue adjacent to the transection site (p = 0.4660) (Fig. 6c, d).
lgmn mutants have an altered immune response to injury
To assess immune cell response in lgmn mutants, we first quantified neutrophils at 1 and 3 dplt using immunofluorescence for the neutrophil marker, Mpx (Supplementary Fig. 4a). Compared to uninjured controls, neutrophils are recruited to the site of ligament injury in both wild type and lgmn mutants. However, we observed a significant increase in the number of neutrophils present at the injury site of lgmn mutants compared to wild types at 1 dplt (p = 0.0213) (Supplementary Fig. 5b). By 3 dplt, we observed a decrease in neutrophils at the injury site to similar levels in both wild type and lgmn mutants (Supplementary Fig. 5b). These data suggest that neutrophil recruitment is transiently heightened in the lgmn mutant injury response.
Next, we characterized the macrophage response in lgmn mutants. At 1 dplt, macrophages home to the injury site, surround the injured ligament tissue, and express fn1a and lgmn during wild type regeneration. At 1 dplt, we found that lgmn mutants had similar numbers of mpeg1.1+ cells present and localized to the expected injury site compared to wild types (Fig. 7a, b). This suggests the dysregulated healing observed in lgmn mutants is not due to an absence of macrophages after transection. Next, to assess for shifts in proportions of macrophage subsets present in lgmn mutants, we quantified the proportion of mpeg1.1+ macrophages expressing fn1a and/or lgmn that were most proximal to the injured tissue (Fig. 7a–c, white dashed line in c represents ROI). Compared to wild types, we observe that the injury site in lgmn mutants is populated by similar proportions of heterogeneous macrophage subsets, with a slight reduction in lgmn single-positive macrophages (Fig. 7c, Supplementary Fig. 6).
a Representative images of smFISH staining using mpeg1.1 (magenta), fn1a (cyan), and lgmn (yellow) probes in IOM ligaments in wild type and lgmn mutants from uninjured and 1 dplt. b Number of mpeg1.1+ macrophages at 1 dplt normalized to ROI area in wild type and lgmn mutants. c Proportion of mpeg1.1+ cells that express fn1a and lgmn. ROI of area quantified in smFISH images is outlined with white dashed lines (a). d Representative images of smFISH staining using fn1a (cyan) probes in IOM ligaments in wild types and lgmn mutants from uninjured and 3 dplt. e Quantification of smFISH fn1a expression within the IOM ligament and regenerating mesenchyme. Region of interest for quantification is outlined with white dashed lines for the ligament and red dashed lines for regenerative mesenchyme (n = 3-5 per time point); error bar represents SEM, p = 0.0003. Scale bars = 100 μm (a, d).
Furthermore, we visualized the relationship between macrophages and proliferating fibroblasts at the injury site using immunofluorescence. Wild types and lgmn mutants with Tg(mpeg1.1:eGFP) were immunostained for eGFP and PCNA. At 1 dplt, we observed that ligamentocytes within the ligament stub re-enter the cell cycle without coincident macrophages (Supplementary Fig. 7, red arrowhead). By 3 dplt in wild types, we find that some proliferating cells are in direct contact with macrophages at the edge of the ligament stub, as well as throughout the mesenchyme domain (Supplementary Fig. 6a–c). However, we observe proliferating cells without coincident macrophages in both the ligament and regenerating mesenchyme domains. In lgmn mutants, while we observe similar numbers of mpeg1.1:eGFP+ cells as in wild type, we see a decrease in PCNA+ cells in the mesenchyme domain (Fig. 6b, Supplementary Fig. 7d). These data suggest that initial ligamentocyte cell cycle re-entry may be independent of crosstalk with macrophages and that lgmn is not required for macrophage homing to the injury site during ligament regeneration.
ECM gene expression changes in the regenerative mesenchyme of lgmn mutants
Given the disorganization of the regenerated ligaments in lgmn mutants, we next assessed potential changes in ECM gene expression. In the uninjured IOM ligament of adult wild types and lgmn mutants, we detected very low expression of fn1a (Fig. 7d). At 1 dplt, we observed upregulation of fn1a expression in wild type and lgmn mutant macrophages, regenerative mesenchyme, and ligament tissue adjacent to the transection site. By 3 dplt, fn1a expression is spatially restricted in wild types, with highest expression at the transection site of the injured ligament and low expression throughout the adjacent regenerative mesenchyme domain (Fig. 7d). In lgmn mutants, we observe a loss of this spatially restricted expression of fn1a. Instead, we observe a significant increase in fn1a expression as well as an expansion of its expression domain in the ligament tissue and throughout the regenerative mesenchyme domain (Fig. 7d, e). Taken together, our data show that reduced mesenchymal proliferation and aberrant ECM gene expression are associated with the failure to properly regenerate the IOM ligament in lgmn mutants.
Discussion
Here we show that the neural crest-derived IOM ligament in adult zebrafish can fully regenerate in one month following complete transection. Lineage tracing experiments show that regeneration is mediated in part by the dedifferentiation, proliferative expansion, and redifferentiation of mature ligamentocytes flanking the injury site. Single-cell expression profiling further reveals dynamic changes in ECM composition during the regenerative process, mediated by both neural crest-derived mesenchyme and immune cells that home to the injury site. In particular, we identify Legumain as a key macrophage-secreted protease required for ECM remodeling, mesenchymal proliferation, and proper ligament regeneration (Fig. 8). These findings provide a paradigm for how cross-talk between the immune system and mesenchyme can promote scar-free regeneration of adult connective tissue.
Zebrafish have emerged as a high-throughput model organism used in understanding the cellular and molecular mechanisms underlying the regeneration of diverse adult vertebrate organs13,15,30. Our zebrafish adult ligament regeneration model differs from mammalian connective tissue regeneration models, such as the neonatal Achilles tendon regeneration model in which tenocytes have retained proliferative potential from development8, and the periodontal ligament regeneration model, in which regeneration relies on a tooth-associated resident stem cell population31. In our model, we show that post-mitotic ligamentocytes in the adult jaw can dedifferentiate and re-enter the cell cycle after injury to fully regenerate the ligament. The concept of mature cells undergoing dedifferentiation and proliferation appears to be a common theme for adult zebrafish regeneration, as seen in cardiomyocytes during heart regeneration32,33 and in osteoblasts during fin bone regeneration34. However, as de novo osteoblasts also contribute to fin bone regeneration35, we cannot rule out that resident stem/progenitor cells may also contribute to ligament regeneration.
Our single-cell expression profiling reveals key changes in ECM composition in early stages of ligament regeneration. Coincident with loss of mature markers of ligamentocytes within ligament tissue adjacent to the transection site, histology reveals a rapid loss of the characteristic ligament ECM in a few days after injury. An intriguing possibility is that initial ECM remodeling facilitates the release and dedifferentiation of ligamentocytes to regenerate the ligament. At the same time, the regenerative mesenchyme bridge transiently acquires a distinct ECM signature including high levels of Type XII Collagen and Fibronectin 1, which is subsequently lost as ligamentocytes differentiate anew. These regenerative mesenchymal cells are also transcriptionally distinct from the αSMA-secreting myofibroblasts which comprise mammalian scar tissue36,37. Interestingly, fibroblasts expressing Type XII Collagen and Fibronectin 1 have also been implicated in heart and spinal cord regeneration in zebrafish, suggesting that a common ECM signature may facilitate scar-free repair across diverse types of organs25,38.
A key modulator of regeneration is the immune system. Neutrophils and macrophages have been shown to be essential for fin, spinal cord, and heart regeneration in zebrafish39,40,41,42. Here we used single-cell expression profiling and in vivo imaging to reveal successive waves of neutrophil and macrophage recruitment to the injury site, as well as injury-induced transcriptional changes in macrophages that accompany ligament regeneration. Macrophages strongly upregulate Fibronectin 1 expression during early phases of ligament repair, and an analogous role for macrophages in collagen deposition has been described for heart regeneration40. Thus, mesenchymal and immune cells may cooperate to build an ECM environment conducive to regeneration.
We found lgmn to be strongly expressed in macrophages recruited to the injured ligament, with loss of lgmn preventing proper ligament regeneration. Whereas Legumain has been shown to be critical for the resolution of fibrosis after murine myocardial infarction and obstructive nephropathy43,44, its role in regeneration had not been previously explored. In zebrafish lgmn mutants, we observed a delay in resorbing ligament tissue immediately adjacent to the injury site and a subsequent failure to regenerate an integrated and properly aligned ligament. These histological defects were accompanied by decreased mesenchymal proliferation and a failure to regulate the spatially restricted expression of fn1a during later phases of regeneration. We found that mpeg1.1+ macrophages and col12a1a+ regenerative mesenchyme were sources of fn1a expression in both wild types and lgmn mutants. Our finding that lgmn mutants have increased numbers of neutrophils present in the injury site at 1 dplt could be due to the recruitment of a larger wave of neutrophils after injury. Alternatively, dysregulated crosstalk between neutrophils and macrophages in lgmn mutants throughout the course of healing could drive abnormal persistence of neutrophils. Our investigation into the role of macrophages in regulating proliferation revealed that initial ligamentocyte cell-cycle re-entry is independent of macrophage crosstalk in both wild types and lgmn mutants. In contrast, some of the later proliferating cells in the regenerating mesenchyme domain were in close contact with macrophages. Due to the highly motile nature of macrophages, we cannot define a direct relationship between macrophages and all proliferating fibroblasts in histological sections. The decreased levels of proliferation in the mesenchyme domain in lgmn mutants indicate that cross-talk between immune cells and mesenchymal cells is potentially important to regulate proliferation of mesenchymal progenitors for ligament regeneration.
The targets of the Legumain protease in our system remain unknown. Legumain is known to cleave and activate Mmp-2, which belongs to the family of metalloproteases known to break down collagen ECM20. One possibility then is that Legumain may function to initiate resorption of adjacent ligament tissue, thus releasing ligamentocytes to participate in the repair process. Legumain has also been shown to degrade fibronectin45,46, and thus could function later to break down the transient fibronectin-rich ECM of the regenerative mesenchyme bridge, in combination with negative regulation of fn1a expression. It is also possible that Legumain works in a protease-independent manner through activation of integrin αvβ347,48 or αvβ149. Future work will be needed to determine the relationship of ECM remodeling to transcriptional and proliferative changes of ligamentocytes and mesenchymal cells. It will also be interesting to assess potential roles of Legumain on immune cells themselves, as well as the extent to which Legumain is required in macrophages versus other cell types. In the future, a comparison of scar-free ligament regeneration in zebrafish with imperfect healing in mammals may reveal new cellular and molecular targets for therapies aimed at alleviating the substantial clinical and societal burden posed by ligament injuries.
Methods
Zebrafish lines
The Institutional Animal Care and Use Committees of Columbia University and the University of Southern California approved all zebrafish experiments. Published Zebrafish lines used in this study include AB, Tübingen, lgmnsa2235050, Tg(scxa:mCherry)51, Tg(actab2:loxP-BFP-STOP-loxP-dsRed)sd2752, Tg(Mmu.Sox10-Mmu.Fos:Cre)zf38453, Tg(mpx:mCherry)uwm754, and Tg(mpeg1:EGFP)gl2255. Embryos were raised in a methylene blue salt solution at 28.5 °C. Juvenile and adult fish were housed in groups of 15-30 individuals. All surgical injuries were performed in size-matched 3-6 months post fertilization (mpf) adult zebrafish using standard body length (SL) measurements.
lgmnsa22350 mutant allele containing a G > T point mutation resulting in a premature stop codon was used to generate homozygous lgmn mutants (referred to as lgmnMUT). Zebrafish were genotyped by PCR with primers flanking intron 1 (lgmn forward primer: AGATCTTATGATCCCAGATCCAATGACT) and end of exon 2 (lgmn reverse primer: GGTGAGAAAATGAAACCCGAAACTAGTCT). The 500 bp PCR product was digested with NspI (New England Biolabs, R0602L) that recognizes a RCATG/Y cut site only present in mutants. Mutant alleles were then identified by presence of the cleaved 358 bp and 142 bp bands.
Generation of thbs4a_p1 transgenic zebrafish lines
The Tg(thbs4a_p1:CreERT2)el913 and Tg(thbs4a_p1:eGFP)el912 alleles were generated using Gateway cloning and Tol2 transgenesis. p5E plasmid including the 635 bp thbs4a_p1 enhancer and minimal E1B promoter sequence was generated using synthesized gBlock DNA (IDT) in a BP cloning reaction (see Supplementary Methods 1 for thbs4a_p1 gBlock sequence). The thbs4a_p1:eGFP transgene was generated by combining p5E-thbs4a_p1:E1B, pME-eGFP, p3E-polyA, and pDestTol2AB2. The thbs4a_p1:CreERT2 transgene was generated by combining p5E-thbs4a_p1:E1B, pME-CreERT2, p3E-polyA, and pDestTol2CG2. Transgenes were injected into one-cell stage zebrafish embryos at 30 ng/ul plasmid DNA with 50 ng/ul Tol2 mRNA. Injected F0 animals were raised to adulthood and outcrossed to Tübingen or Tg(actab2:loxP-BFP-STOP-loxP-dsRed)sd27 animals. F1 founders were screened for the presence of eye CFP (thbs4a_p1:eGFP) or heart GFP and successful 4-OHT conversion in progeny (thbs4a_p1:CreERT2), respectively.
Adult ligament transection surgery
Interopercular-mandibular (IOM) ligament transection surgery was adapted from Smeeton et al.18. Briefly, adult zebrafish were anesthetized with Tricaine MS-222 and restrained in a damp sponge. Using 3-mm Vannas spring scissors (Fine Science Tools, cat. #1500000), the IOM ligament was severed with a single cut and a pull test performed on the IOP bone to confirm transection. Fish were then revived in clean system water, housed on-system, and received daily post-operative health checks for three days. At experimental endpoints, zebrafish were euthanized using a lethal dose of Tricaine MS-222 with rapid chilling on ice.
Histology, in situ hybridization, and immunohistochemistry
Adult zebrafish samples were fixed in 4% PFA at 4 °C overnight. Following fixation, samples were washed twice for 30 minutes in 1xPBS and cut to facilitate embedding and sectioning of the head. Samples were decalcified in 0.5 M EDTA solution for 10 days rocking at room temperature. For embedding, the tissue was dehydrated in 20 min increments with a series of ethanol dilutions (30, 50, 70, 95, and 100%). The water was then replaced with Hemo-De Xylene Substitute (Electron Microscopy Sciences, 23412-01) in a series (50% Hemo-De/EtOH, 75% Hemo-De/EtOH, 100% Hemo-De) of 15 min washes. Then the samples were incubated in a 1:1 Hemo-De:Paraffin solution at 65 °C for an hour before overnight incubation in 100% paraffin at 65 °C. Samples were then embedded in freshly melted paraffin. Paraffin sectioning was performed using Thermo HM355S automatic microtome to collect 5 μm sections.
For AFOG staining, 5μm paraffin sections were deparaffinized with Hemo-De and re-hydrated through an ethanol series (100, 90, 70, 50, 0) diluted with water. Sections were then fixed in 4% PFA for 5 min at RT, then washed 3x for 5 min with 1xPBS and then washed 1x for 5 min in water. Following washes, samples were incubated in Bouin’s solution (Sigma-Aldrich, HT10132) that was preheated to 60 °C for 2-hours and cooled to room temperature for another hour. Slides were then washed 5-6x for 5 min with water and then rinsed in 1% Phosphomolybdic acid (Sigma-Aldrich, HT153) for 10 min and rinsed in water for 5 min. Slides were then incubated in AFOG staining solution (0.5% aniline blue, 1% orange-G, and 1.5% acid fuchsin, pH 1.09; Sigma-Aldrich, 415049, O3756, F8129) for 7 min at RT and then rinsed 1x for 2 min with water. Samples were dehydrated with a series of ethanol dilutions (75, 90, 100%) and 1x in Hemo-De for 5 min before mounted with Cytoseal (Epredia, 23-244256).
For Toluidine Blue/Fast Green staining, zebrafish tissue was fixed and paraffin-embedded according to the aforementioned paraffin embedding protocol. Paraffin blocks were sectioned using HM 355 S automated microtome and MX35 microtome blades at 5μm. Sections were floated in DEPC-treated water (Sigma-Aldrich, D5758) in a waterbath and collected onto Leica adhesive slides, then allowed to air dry at room temperature at least overnight. Slides were then processed for toluidine blue staining with the following procedure: two 5-minute washes in xylene substitute Hemo-De, two 1-minute washes in 100% ethanol; serial washes (90%, 70%, and 50%) in EtOH/H2O for 1 minute per wash; and three 1-minute washes in tap water. Slides were then placed in 0.04% toluidine blue solution (0.08 g toluidine blue in 200 mL of 0.1 M sodium acetate, pH 4.0; Sigma-Aldrich, 89640) for 10 minutes, followed by three 1-minute washes in tap water. Slides were then placed in 0.1% fast green solution (0.2 g fast green in 200 mL water; Fisher Scientific, BP123-10) for 3 minutes, followed by three 1-minute washes in tap water. Stained slides were then given two 1-minute washes in 100% isopropanol and two 1-minute washes in Hemo-De. Lastly, coverslips were mounted on the slides using CytoSeal and allowed to air dry for 30 minutes to overnight.
Tissue clearing
CUBIC tissue clearing protocol was adapted from (Susaki et al. 56) for use with zebrafish. Whole adult zebrafish were fixed overnight in the dark at 4 °C in 4% PFA. Tissue was then washed twice in PBS for at least 30 minutes per wash and heads were separated from the trunk of the body. At a ratio of 5 zebrafish heads per 50 mL of solution, tissue was incubated in the dark at 37 °C in CUBIC-1 solution with gentle rocking for at least 3 days. Tissue was then transferred to new CUBIC-1 to incubate until tissue was transparent and iridophores were cleared (2-4 days). Tissue was then washed twice in PBS for at least 30 minutes per wash, then immersed in 20% sucrose in PBS at room temperature until tissue was no longer floating in solution and once again opaque, 2 hours to overnight. Lastly, the tissue was immersed in CUBIC-2 solution and incubated in the dark at room temperature with gentle rocking for at least 1 day before imaging. Fluorescent microscopy images were captured using a Leica SP8 confocal microscope and processed in Fiji. CUBIC-1 was prepared as a mixture of 25 wt% urea, 25 wt% N,N,N’,N’-tetrakis(2-hydroxypropyl) ethylenediamine (Fisher Scientific, AAL16280AP) and 15 wt% Triton X-100. To minimize water evaporation, solution was prepared with heat not exceeding 100 °C and inside bottles with caps just screwed on. Deionized water was heated prior to the addition of urea. Once urea was dissolved and the solution was no longer cold, N,N,N’,N’-Tetrakis(2-hydroxypropyl)ethylenediamine was mixed until homogenous. Solution was removed from heat and allowed to cool to room temperature, then Triton-X-100 was added with gentle stirring to minimize the production of bubbles. CUBIC-1 solution was stored in the dark at room temperature and used within 1 week.
CUBIC-2 was prepared as a mixture of 50 wt% sucrose, 25 wt% urea, 10 wt% 2,2′,2′’-nitrilotriethanol (Fisher Scientific, AAL044860E), and 0.1% (v/v) Triton X-100. To minimize water evaporation, solution was prepared with heat not exceeding 100 °C and inside bottles with caps just screwed on. Deionized water was heated prior to the addition of urea. Once urea was dissolved and the solution was no longer cold, 2,2′,2′′-Nitrilotriethanol was mixed until homogenous. Sucrose was mixed with continued heat until the solution was clear with a pale yellow tint. Solution was removed from heat and allowed to cool to room temperature, then Triton-X-100 was added with gentle stirring to minimize the production of bubbles. CUBIC-2 solution was stored in the dark at room temperature and used within 1 week.
Immunofluorescence and smFISH
We collected 5μm formalin-fixed paraffin-embedded (FFPE) tissue sections. For immunofluorescence, sections were deparaffinized with Hemo-De and rehydrated through an ethanol series, then subjected to antigen retrieval in sodium citrate buffer, blocked with Agilent Dako protein block (cat. X090930-2), and incubated with primary antibody (PCNA, 1:1000; Sigma-Aldrich, P8825) (mCherry [used to detect DsRed], 1:200; Novus, NBP2-25157SS) (Mpx, 1:200; GeneTex, GTX128379), (eGFP 1:200; OriGene, TP401) overnight at 4 °C, washed, and incubated with secondary antibody (1:500; Invitrogen, PIA32727). TUNEL staining was performed as per manufacturer’s instructions using the Apoptag Fluorescein In Situ Apoptosis Detection Kit (Millipore Sigma, S7110). Slides were mounted and counterstained with Fluoromount-G + DAPI (Southern Biotech, 0100-20). Single-molecule fluorescence in situ hybridization was performed on 5μm FFPE sections according to manufacturer guidelines using RNAScope Multiplex Fluorescent Reagent Kit v2 Assay (ACD Bio). Slides were treated with 1x Target Retrieval Reagent for 4 min in a steamer. RNAScope probes used were Dr-acta2 (508581-C4), Dr-col12a1a (556481-C2), DsRed-C2 (481361-C2), Dr-fn1a (1097911-C1), Dr-scxa (564451), Dr-lgmn (1003381-C2), Dr-mpeg1.1 (536171-C3), and Dr-thbs4a (812151) from ACD Bio. Opal 520, 570, and 690 fluorophores were used to visualize expression (1:1,000; Akoya Biosciences, cat. #FP1487001KT, #FP1488001KT, and #FP1497001KT). Sections were counterstained and mounted with Fluoromount-G + DAPI.
Drug treatments
Adult thbs4a_p1:CreERT2 zebrafish lines were converted with three 5 µM (Z)-4-Hydroxytamoxifen, ≥98% Z isomer (Sigma-Aldrich, H7904) overnight treatments in the dark. Following drug treatment, adults were screened for conversion using Leica M165FC stereo microscope and then washed 2 ×1 hour in system water before being re-housed on-system.
Single-cell RNA sequencing
Ligament transection (n = 22 joints per 1 dplt and 3 dplt time points), sham surgery (n = 22 joints per 1 d and 3 d time point) was performed on size-matched Sox10Cre;actb2:loxP-BFP-loxP-DsRed fish at 3-4 mpf. The jaw joint region of each fish and uninjured control (25 joints) was microdissected and incubated in protease solution [0.25% trypsin (Life Technologies, 15090–046), 1 mM EDTA, and 400 mg/ml collagenase D (Sigma-Aldrich, 11088882001) in PBS] with mechanical dissociation by nutation and agitation with a P1000 pipette every 5 min for 1 h at 28 °C. Cells were FACS sorted and immediately processed to create single-cell RNA Sequencing libraries using the Chromium Single Cell 3′ Library & Gel Bead Kit v2 (10X Genomics), according to the manufacturer’s recommendations. To determine cell count, libraries were sequenced at the CHLA Center for Personalized Medicine Genomics Core using a MiSeq Nano v2 (Illumina, San Diego, CA) with 26 bp sequencing for Read1, 8 bp sequencing for Index1, and 120 bp sequencing for Read2. Libraries were pooled for even coverage per cell and were sequenced at the CHLA Center for Personalized Medicine Molecular Genomics Core on the NextSeq 550 (Illumina, San Diego, CA) with 26 bp sequencing for Read1, 8 bp sequencing for Index1, and 120 bp sequencing for Read2. All samples were sequenced to an average read depth of greater than 130,000 reads per cell. CellRanger v3.0.0 (10X Genomics) was used with default parameters for alignment to GRCz11 to generate gene-by-cell count matrices.
Analyses of scRNAseq libraries were performed using R version 4.0.5 in RStudio with Seurat version 4.0.2. CellRanger output for all samples were converted to Seurat objects and merged, then filtered for cells with between 400 and 1,900 unique features per cell, with under 5% mitochondrial RNA detected per cell to remove low-quality cells, doublets, or dying cells. Data were log-normalized before selecting 2,000 variable features, then scaled using all features expressed in at least 3 cells. Linear dimensional reduction was then performed using principal component analysis to compute 80 principal components. Statistical significance for each principal component was determined with a jackstraw permutation, which identified 68 significant principal components (p < 0.05). Principal components 1-68 were used for K-nearest-neighbor graph construction (KNN). Clustering was performed at low resolution (r = 0.02) to identify broad superclusters for downstream analysis (Fig. 2a, b). Clusters and gene expression were graphed using UMAP-non-linear dimensional reduction to preserve global variability.
Analysis of scRNAseq data from neural crest-derived cells were performed by subsetting and merging the superclusters containing cells from the DsRed-enriched samples (Fig. 3c). Using the unprocessed data from these cells, gene expression data were again processed with log-normalization, feature selection, and feature scaling. Linear dimensional reduction was then performed using principal component analysis to identify 50 principal components, of which 38 were deemed significant using a jackstraw (p < 0.05). Principal components 1-38 were used for KNN and graphed using UMAP-non-linear dimensional reduction before clustering at r = 0.95. Cluster 13, which showed uncharacteristically high unique feature counts and markers for epithelial cells (epcam, cdh1), were excluded as a likely doublet cluster. Markers for each cluster were identified with function FindAllMarkers using a Wilcoxon Rank Sum test for features expressed in at least 10% of the cluster and a minimum of 0.2 log-fold-change higher expression than the other cells in the neural crest-derived superclusters.
Analysis of scRNAseq data from immune cells were performed by subsetting and merging the superclusters Immune 1 and Immune 2 (Fig. 3b). Using the unprocessed data from these cells, gene expression data were again processed with log-normalization, feature selection, and feature scaling. Principal components 1-50 were used for KNN and graphed using UMAP-non-linear dimensional reduction before clustering at r = 0.9. Markers for each cluster were identified with the function FindAllMarkers using a Wilcoxon Rank Sum test for features expressed in at least 10% of the cluster and a minimum of 0.25 log-fold-change higher expression than the other cells in the immune superclusters.
Imaging
Images of histological sections were captured using a Keyence BZ-X810 microscope and software, then processed in Fiji. Live imaging was performed using a Leica M165FC stereo microscope with Leica K5 sCMOS camera and LASX software v3.7.4.23463. Fluorescence imaging was performed using a Leica SP8 confocal microscope with LASX software v3.5.7.23225. The 3-D projections of mpeg1.1:eGFP+ macrophages and PCNA+ cells were generated from z-stacks obtained from 63x confocal imaging using the LASX-3D software v3.5.7.23225.
Quantification and statistical analysis
Proliferation was quantified as a percentage PCNA+ cells of total DAPI+ cells in a region of interest (ROI) in the uninjured ligament domain, ligament stub domain, or regenerative mesenchyme domain and scored by an observer blinded to treatment and/or genotype. Wild type and lgmn mutant PCNA staining and quantification were performed concurrently and wild type quantification is shown both in Fig. 2f and as wild-type controls in Fig. 6d. ImageJ was used to quantify fn1a expression levels from smFISH images. Ligament domains and mesenchyme domains were drawn based on regional nuclear morphology in the tissue section. To calculate the corrected total fluorescence, an ROI was drawn to measure the integrated density for each channel using the same ROI for each channel. To account for tissue autofluorescence and background signal, 3 background regions of each image were measured, and the average mean gray value was calculated. To calculate the corrected total fluorescence (CTF), we used the following formula CTF = Integrated Density-(area of ROI*background mean). ImageJ was used on smFISH images to quantify the proportion of mpeg1.1+ cells that were also positive for fn1a and/or lgmn that surrounded the ligament stub by a blinded observer. The ROI for quantification was drawn to include 2-3 cell layers most proximal to the ligament stub excluding skin cells. ImageJ was used to quantify the number of Mpx+ cells by a blinded observer. The region of interest for quantification of Mpx+ cells was drawn to include the entire injury area.
All statistical analysis was carried out using GraphPad Prism 9. Proliferation in wild type animals was performed by ordinary 1-way ANOVA with Tukey’s multiple comparison test. Comparison of wild type to lgmn mutant proliferation, TUNEL, and fn1a transcript expression were each performed by 2-way ANOVA with Šídák’s multiple comparisons test. Comparison of wild type to lgmn mutant Mpx+/ROI was performed by 2-way ANOVA with Tukey’s multiple comparisons test. Comparison of wild type and lgmn mutant macrophage expression of fn1a and/or lgmn, and mpeg1.1:eGFP+ cell abundance were performed with Student’s T-test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Raw and processed scRNA sequencing files have been deposited in GEO and can be retrieved using accession number GSE224197. All other raw data, transgenic zebrafish lines, and/or materials from this study are available from the corresponding author (JS) upon request.
Code availability
Code to analyze single-cell sequencing data used publicly available software (R v4.0.5, Seurat v4.0.2, dplyr v1.0.10. ggplot v3.3.6, Matrix v1.5-3, patchwork v1.1.2) and is available from the corresponding author (JS) upon request.
References
Gelber, A. C. et al. Joint injury in young adults and risk for subsequent knee and hip osteoarthritis. Ann. Intern. Med. 133, 321–328 (2000).
Lohmander, L. S., Östenberg, A., Englund, M. & Roos, H. High prevalence of knee osteoarthritis, pain, and functional limitations in female soccer players twelve years after anterior cruciate ligament injury. Arthritis Rheum. 50, 3145–3152 (2004).
Altman, R. D., Kates, J., Chun, L. E., Dean, D. D. & Eyre, D. Preliminary observations of chondral abrasion in a canine model. Ann. Rheum. Dis. 51, 1056–1062 (1992).
Wright, R. W. et al. Effect of hemorrhage on medial collateral ligament healing in a mouse model. Am. J. Sports Med. 31, 660–666 (2003).
Shrive, N. et al. Soft-tissue “flaws” are associated with the material properties of the healing rabbit medial collateral ligament. J. Orthop. Res. 13, 923–929 (1995).
Frank, C. et al. Medial collateral ligament healing: A multidisciplinary assessment in rabbits. Am. J. Sports Med. 11, 379–389 (1983).
Bray, R. C., Rangayyan, R. M. & Frank, C. B. Normal and healing ligament vascularity: a quantitative histological assessment in the adult rabbit medial collateral ligament. J. Anat. 188, 87–95 (1996).
Howell, K. et al. Novel model of tendon regeneration reveals distinct cell mechanisms underlying regenerative and fibrotic tendon healing. Sci. Rep. 7, 45238 (2017).
Murray, M. M., Martin, S. D., Martin, T. L. & Spector, M. Histological changes in the human anterior cruciate ligament after rupture*. JBJS 82, 1387 (2000).
Nguyen, D. T. et al. Intrinsic healing response of the human anterior cruciate ligament: An histological study of reattached ACL remnants. J. Orthop. Res. 32, 296–301 (2014).
Maier, D. et al. The acromioclavicular ligament shows an early and dynamic healing response following acute traumatic rupture. BMC Musculoskelet. Disord. 21, 593 (2020).
Niu, X., Subramanian, A., Hwang, T. H., Schilling, T. F. & Galloway, J. L. Tendon cell regeneration is mediated by attachment site-resident progenitors and BMP signaling. Curr. Biol. 30, 3277–3292.e5 (2020).
Poss, K. D., Wilson, L. G. & Keating, M. T. Heart regeneration in zebrafish. Science 298, 2188–2190 (2002).
Becker, T., Wullimann, M. F., Becker, C. G., Bernhardt, R. R. & Schachner, M. Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 377, 577–595 (1997).
Géraudie, J., Brulfert, A., Monnot, M. J. & Ferretti, P. Teratogenic and morphogenetic effects of retinoic acid on the regenerating pectoral fin in zebrafish. J. Exp. Zool. 269, 12–22 (1994).
Askary, A. et al. Ancient origin of lubricated joints in bony vertebrates. eLife 5, e16415 (2016).
Smeeton, J., Askary, A. & Crump, J. G. Building and maintaining joints by exquisite local control of cell fate. WIREs Dev. Biol. 6, e245 (2017).
Smeeton, J. et al. Regeneration of jaw joint cartilage in adult zebrafish. Front. Cell Dev. Biol. 9, 777787 (2022).
Freeley, S. et al. Asparaginyl endopeptidase (Legumain) supports human Th1 induction via cathepsin L-mediated intracellular C3 Activation. Front. Immunol. 9, 2449 (2018).
Bai, P. et al. Macrophage-derived legumain promotes pulmonary hypertension by activating the MMP (Matrix Metalloproteinase)-2/TGF (Transforming Growth Factor)-β1 Signaling. Arterioscler. Thromb. Vasc. Biol. 39, e130–e145 (2019).
Schweitzer, R. et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855–3866 (2001).
Pryce, B. A., Brent, A. E., Murchison, N. D., Tabin, C. J. & Schweitzer, R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev. Dyn. 236, 1677–1682 (2007).
Espira, L. et al. The basic helix–loop–helix transcription factor scleraxis regulates fibroblast collagen synthesis. J. Mol. Cell. Cardiol. 47, 188–195 (2009).
Fabian, P. et al. Lifelong single-cell profiling of cranial neural crest diversification in zebrafish. Nat. Commun. 13, 13 (2022).
Wehner, D. et al. Wnt signaling controls pro-regenerative Collagen XII in functional spinal cord regeneration in zebrafish. Nat. Commun. 8, 126 (2017).
Xia, Y. et al. Activation of a transient progenitor state in the epicardium is required for zebrafish heart regeneration. Nat. Commun. 13, 7704 (2022).
Hu, B. et al. Origin and function of activated fibroblast states during zebrafish heart regeneration. Nat. Genet. 54, 1227–1237 (2022).
Tsata, V. et al. A switch in pdgfrb+ cell-derived ECM composition prevents inhibitory scarring and promotes axon regeneration in the zebrafish spinal cord. Dev. Cell 56, 509–524.e9 (2021).
Carmona, S. J. et al. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 27, 451–461 (2017).
Becker, C. G. & Becker, T. Adult zebrafish as a model for successful central nervous system regeneration. Restor. Neurol. Neurosci. 26, 71–80 (2008).
Seo, B.-M. et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 364, 149–155 (2004).
Jopling, C. et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609 (2010).
Kikuchi, K. et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601–605 (2010).
Knopf, F. et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713–724 (2011).
Singh, S. P., Holdway, J. E. & Poss, K. D. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 22, 879–886 (2012).
Faryniarz, D. A., Chaponnier, C., Gabbiani, G., Yannas, I. V. & Spector, M. Myofibroblasts in the healing lapine medial collateral ligament: Possible mechanisms of contraction. J. Orthop. Res. 14, 228–237 (1996).
Darby, I., Skalli, O. & Gabbiani, G. Alpha-smooth muscle actin is transiently expressed by myofibroblasts during experimental wound healing. Lab. Investig. J. Tech. Methods Pathol. 63, 21–29 (1990).
Marro, J. et al. Collagen XII contributes to epicardial and connective tissues in the zebrafish heart during ontogenesis and regeneration. PLOS ONE 11, e0165497 (2016).
Petrie, T. A., Strand, N. S., Tsung-Yang, C., Rabinowitz, J. S. & Moon, R. T. Macrophages modulate adult zebrafish tail fin regeneration. Development 141, 2581–2591 (2014).
Simões, F. C. et al. Macrophages directly contribute collagen to scar formation during zebrafish heart regeneration and mouse heart repair. Nat. Commun. 11, 600 (2020).
Bevan, L. et al. Specific macrophage populations promote both cardiac scar deposition and subsequent resolution in adult zebrafish. Cardiovasc. Res. 116, 1357–1371 (2020).
Tsarouchas, T. M. et al. Dynamic control of proinflammatory cytokines Il-1β and Tnf-α by macrophages in zebrafish spinal cord regeneration. Nat. Commun. 9, 4670 (2018).
Jia, D. et al. Cardiac resident macrophage-derived legumain improves cardiac repair by promoting clearance and degradation of apoptotic cardiomyocytes after myocardial infarction. Circulation 145, 1542–1556 (2022).
Wang, D. et al. Legumain, an asparaginyl endopeptidase, mediates the effect of M2 macrophages on attenuating renal interstitial fibrosis in obstructive nephropathy. Kidney Int. 94, 91–101 (2018).
Morita, Y. et al. Legumain/asparaginyl endopeptidase controls extracellular matrix remodeling through the degradation of fibronectin in mouse renal proximal tubular cells. FEBS Lett. 581, 1417–1424 (2007).
Jafari, A. et al. Legumain regulates differentiation fate of human bone marrow stromal cells and is altered in postmenopausal osteoporosis. Stem Cell Rep. 8, 373–386 (2017).
Pan, L. et al. Legumain is an endogenous modulator of integrin αvβ3 triggering vascular degeneration, dissection, and rupture. Circulation 145, 659–674 (2022).
Liu, Y., Bajjuri, K. M., Liu, C. & Sinha, S. C. Targeting cell surface alpha(v)beta(3) integrin increases therapeutic efficacies of a legumain protease-activated auristatin prodrug. Mol. Pharm. 9, 168–175 (2012).
Li, X. et al. The exosomal integrin α5β1/AEP complex derived from epithelial ovarian cancer cells promotes peritoneal metastasis through regulating mesothelial cell proliferation and migration. Cell. Oncol. 43, 263–277 (2020).
Busch-Nentwich, E. et al. Sanger Institute Zebrafish Mutation Project mutant data submission. (2013).
McGurk, P. D., Swartz, M. E., Chen, J. W., Galloway, J. L. & Eberhart, J. K. In vivo zebrafish morphogenesis shows Cyp26b1 promotes tendon condensation and musculoskeletal patterning in the embryonic jaw. PLOS Genet 13, e1007112 (2017).
Kobayashi, I. et al. Jam1a–Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 512, 319–323 (2014).
Kague, E. et al. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLOS ONE 7, e47394 (2012).
Yoo, S. K. et al. Differential regulation of protrusion and polarity by PI(3)K during neutrophil motility in live zebrafish. Dev. Cell 18, 226–236 (2010).
Ellett, F., Pase, L., Hayman, J. W., Andrianopoulos, A. & Lieschke, G. J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Acknowledgements
We thank Joshua Barber, Megan Matsutani and Jennifer DeKoeyer Crump for fish care, Jeffrey Boyd at the USC Flow Core, and Dave Ruble at the CHLA Sequencing Core. Funding was provided by the National Institute of Dental and Craniofacial Research [NIH F31 DE031970 (T.A), R35 DE027550 (J.G.C.) and R00 DE027218 (J.S.)], T32 training programs NIGMS GM007088 (T.A. and J.M.) and GM008224 (M.B.), Columbia Stem Cell Initiative Seed Fund Award (J.S.), Barnard SRI (H.L.), and CIRM training program (K.-C.T.). The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Contributions
T.A. and J.M. contributed equally to this study. J.S. devised the surgical injury model. J.G.C. and J.S. conceived the project. T.A., J.M., E.G., D.S., M.B., E.C., G.L., and J.S. performed the injury experiments and analyses. K.-C.T., P.F., and J.S. performed the scRNAseq and pre-processed the data. J.S. generated and screened the thbs4a_p1:eGFP zebrafish line. J.G.C., H.-J.C., and J.S. generated and screened the thbs4a_p1:CreER zebrafish line. T.A. and J.S. performed the scRNAseq data analysis. T.A., J.M., E.G., and J.S. wrote the first draft of the manuscript with editing by J.G.C. and input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Anderson, T., Mo, J., Gagarin, E. et al. Ligament injury in adult zebrafish triggers ECM remodeling and cell dedifferentiation for scar-free regeneration. npj Regen Med 8, 51 (2023). https://doi.org/10.1038/s41536-023-00329-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41536-023-00329-9