Abstract
Multi-gene panel testing has led to the detection of pathogenic/likely pathogenic (P/LP) variants in many cancer susceptibility genes in patients with breast-ovarian cancer spectrum. However, the clinical and genomic data of Asian populations, including Thai cancer patients, was underrepresented, and the clinical significance of multi-gene panel testing in Thailand remains undetermined. In this study, we collected the clinical and genetic data from 4567 Thai patients with cancer in the hereditary breast-ovarian cancer (HBOC) spectrum who underwent multi-gene panel testing. Six hundred and ten individuals (13.4%) had germline P/LP variants. Detection rates of germline P/LP variants in breast, ovarian, pancreatic, and prostate cancer were 11.8%, 19.8%, 14.0%, and 7.1%, respectively. Non-BRCA gene mutations accounted for 35% of patients with germline P/LP variants. ATM was the most common non-BRCA gene mutation. Four hundred and thirty-two breast cancer patients with germline P/LP variants (80.4%) met the current NCCN genetic testing criteria. The most common indication was early-onset breast cancer. Ten patients harbored double pathogenic variants in this cohort. Our result showed that a significant proportion of non-BRCA P/LP variants were identified in patients with HBOC-related cancers. These findings support the benefit of multi-gene panel testing for inherited cancer susceptibility among Thai HBOC patients. Some modifications of the testing policy may be appropriate for implementation in diverse populations.
Similar content being viewed by others
Introduction
Breast cancer is the most common malignancy worldwide and has contributed to a significant impact on global cancer-related deaths1,2. Hereditary cancer syndromes accounted for ~5–10% of all cancer patients3,4. Breast cancer is among the most common cancers with genetic susceptibility, with BRCA1 and BRCA2 regarded as the most identified genes5. Besides BRCA1/2, many pathogenic/likely pathogenic (P/LP) variants in high and moderate penetrance genes for breast-ovarian cancer, including TP53, CDH1, PALB2, STK11, PTEN, CHEK2, ATM, BARD1, BRIP1, and RAD51D, were increasingly identified after the advent of next-generation sequencing (NGS)-based testing5,6,7. Many studies have shown that non-BRCA cancer susceptibility genes contribute to increased breast and other cancer risk8,9. With expanded access to comprehensive and lower-cost NGS-based multi-gene panels, more patients and families with cancer-predisposing gene mutations could be found, leading to proper screening, early detection, and cancer prevention in at-risk individuals10.
The prevalence of BRCA1 and BRCA2 pathogenic variants in breast and other related cancers in various populations and clinical management guidelines of affected individuals were well established, and similar data of non-BRCA breast cancer susceptibility genes were also increasingly published. Though most prevalence data of non-BRCA pathogenic variants in hereditary breast cancer were from Western countries, several studies in the Asian population have shown the significant detection rate and distribution of pathogenic variants in non-BRCA genes in different countries11,12,13,14,15. A study by Su Y et al.11 in China identified 12.2% of high-risk breast cancer patients who harbored pathogenic variants in non-BRCA genes. Another study in India showed that 15.1% of germline pathogenic variants in breast-ovarian cancer patients were from non-BRCA genes15. These findings have highlighted the importance of expanding genetic tests beyond BRCA1 and BRCA2 for breast and ovarian cancer patients.
Germline BRCA1 and BRCA2 testing for clinically indicated breast and ovarian cancer patients has proven cost-effective worldwide and in Thailand16,17,18. The test is covered by health insurance and integrated into many national healthcare systems worldwide. In Thailand, the universal reimbursement of germline BRCA1 and BRCA2 genetic testing for breast cancer patients has been approved since 2022. However, recommendations regarding genetic testing other than BRCA1 and BRCA2 continue to vary between countries, and the data and testing policy of non-BRCA genes among Thai patients with the hereditary breast-ovarian cancer (HBOC) spectrum remains insufficient. Therefore, we aim to identify the prevalence of hereditary cancer in Thai patients with HBOC spectrum and the contribution of non-BRCA breast cancer susceptibility genes detected by multi-gene panel testing. We also aim to demonstrate the clinical phenotypes of patients with identified pathogenic/likely pathogenic (P/LP) variants and compare those clinical phenotypes to the current testing criteria from the U.S. National Comprehensive Cancer Network (NCCN) clinical practice guidelines.
Results
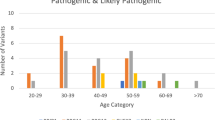
There were 4567 patients with HBOC spectrum tested with multi-gene panel testing due to HBOC-related cancers, consisting of 4041 breast, 394 ovarian (including fallopian tube and primary peritoneal cancer), 100 pancreatic, and 85 prostate cancer patients (Fig. 1). Six hundred and ten patients (13.4%) had at least one P/LP variant. Mutations in BRCA1, BRCA2, and non-BRCA cancer susceptibility genes accounted for 34.9% (n = 213), 31.6% (n = 193), and 35.1% (n = 214), respectively. The detection rate and distribution of P/LP variants in patients with each type of cancer are demonstrated in Table 1.
From 214 cancer patients with non-BRCA P/LP variants, 215 P/LP variants were identified (Fig. 2). ATM was the most commonly identified gene with P/LP variants in 44 individuals (20.5%), followed by PALB2 (n = 38) and TP53 (n = 24). Other mutated genes are categorized in Fig. 2. A summary of patients’ and family history according to each BRCA and non-BRCA variant is demonstrated in Table 2.
Distribution of cancers in patients with P/LP variants
Of 537 breast cancer patients with P/LP variants, P/LP variants in moderate-to-high penetrance breast cancer susceptibility genes were identified in 491 individuals. Meanwhile, there were 78 ovarian, 14 pancreatic, and six prostate cancer patients with P/LP variants. The detection rate of P/LP variants was 19.8% in ovarian cancer patients, followed by pancreatic (14%), breast (11.8%), and prostate cancer (7.1%).
Fifty-four patients had multiple primary cancers, consisting of 27 patients with HBOC-related cancers. The other 27 individuals had additional cancers not in the HBOC spectrum, including colorectal cancer (n = 13), endometrial cancer (n = 11), sarcomas (n = 4) (see Supplementary Table 1). In addition, 162 patients (26.6%) had a history of cancer in their families.
Multi-gene panel testing in breast cancer and NCCN indication fulfillment
Of 491 breast cancer patients with P/LP variants in breast cancer susceptibility genes, 72.3% (n = 355) were BRCA1 or BRCA2 variants, while 139 patients had non-BRCA variants. ATM was the most commonly identified non-BRCA gene (n = 37, 26.6%), followed by PALB2 (n = 36, 25.9%). Details of P/LP variants in other non-BRCA genes are shown in Table 1.
A total of 432 breast cancer patients with P/LP variants (80.4%) fulfilled the 2023 NCCN testing criteria for high-penetrance breast cancer susceptibility genes (Table 3). The most common indication was early-onset breast cancer (n = 365), followed by family history of HBOC-related cancers (n = 102), multiple primary breast cancer (n = 57), triple-negative breast cancer (n = 50), primary breast and ovarian cancers (n = 22), and male breast cancer (n = 10). The proportion of patients with P/LP variants in BRCA and non-BRCA genes who met each criterion is shown in Table 3.
In this cohort, 908 breast cancer patients who received genetic testing did not meet current NCCN criteria. In this group, 105 patients (11.6%) harbored P/LP variants in BRCA1/2 (n = 59) and non-BRCA genes (n = 47). Thirty-two patients with P/LP variants were found in moderate-to-high penetrance non-BRCA genes. Clinical data on patients not meeting the criteria are available in Supplementary Table 2.
Multi-gene panel testing in other HBOC-related cancers
Of 78 ovarian cancer patients with germline P/LP variants, 40 had BRCA1, 17 had BRCA2, and 22 had non-BRCA variants (Table 1). P/LP variants in mismatch repair (MMR) genes (MLH1, MSH2, and PMS2) accounted for 14.4% (n = 11) (see Supplementary Table 3).
There were 14 pancreatic cancer patients with germline P/LP variants, including 6 with BRCA1/2 variants and 8 with non-BRCA variants. Six out of 84 prostate cancer patients also had P/LP variants. Detailed results are shown in Table 1.
Detection of double pathogenic variants in HBOC patients
This cohort identified ten individuals with double pathogenic variants (Table 4). Nine patients had concomitant non-BRCA and BRCA1 or BRCA2 P/LP variants. One patient found both pathogenic RAD51C and CDKN2A variants.
Discussion
This study provides insights into germline mutations and cancer phenotypes in Thai patients with HBOC spectrum and demonstrates the favorable diagnostic yield of multi-gene panel testing in Thai cancer patients. In this study, 13.4% of Thai patients with HBOC-related cancers were associated with cancer susceptibility genes. Among P/LP variants, non-BRCA genes accounted for 35% of these cases. The proportion of non-BRCA variants in breast cancer patients with germline mutation in this study is consistent with previous studies, ranging from 6.8% to 40% in high-risk breast cancer patients11,12,13,14,15,19,20. P/LP variants in non-BRCA genes are identified in 28.3% of breast and 28.2% of ovarian cancer patients with germline mutations. This suggests that the incorporation of additional cancer susceptibility genes in the test for Thai patients with HBOC spectrum may enhance the diagnostic yield by as much as 28% in comparison to BRCA-only testing. Our result aligned with a prior study that supports multi-gene panel testing among breast cancer patients21. Our findings may guide physicians to consider multi-gene panel testing for patients with HBOC-related cancers.
While PALB2 and TP53 were the most reported non-BRCA genes in breast cancer patients in various studies11,12,13,14, ATM was the most common non-BRCA gene mutations in our cohort. From previous studies, the prevalence of ATM P/LP variants among non-BRCA mutated breast cancer patients varied widely11,13,14. A meta-analysis also suggested a pooled prevalence of 7% in P/LP ATM variants among high-risk breast cancer cohorts22. This data may indicate a higher frequency of ATM carriers in the Thai population. ATM mutations are associated with breast and other cancer susceptibilities23,24 and may confer a risk of contralateral breast cancer in patients undergoing radiotherapy25. Despite its relevance, this gene is not included in the NCCN guidelines. Our results support the inclusion of ATM into the breast cancer gene panel for the Thai population.
Our study reveals that only 80% of breast cancer patients with germline mutations met the NCCN criteria for genetic testing. The observation that one-fifth of Thai breast cancer patients with pathogenic variants would miss out on testing opportunities aligned with a previous study in which NCCN criteria missed around 30% of patients with pathogenic variants26. It has been suggested that lowering the age threshold for universal genetic testing could improve the detection rate in breast cancer patients27, which was supported by the 2019 American Society of Breast Surgeons28. Owing to the high acceptance of genetic testing and counseling, more detection of cases would benefit treatment, screening, and prevention for patients and family members carrying pathogenic variants29,30.
The diagnostic yield of germline testing in Thai ovarian cancer patients was 20%, which is comparable with previous studies31,32,33. Mismatch repair (MMR) genes accounted for 14% of P/LP variants in ovarian cancer patients, consistent with the evidence that supports an association between ovarian cancer and Lynch syndrome5,34,35. These findings should raise physicians’ awareness of genetic testing beyond BRCA1/2 and encourage the inclusion of MMR genes in the panel for Thai ovarian cancer patients.
Twenty-seven patients (4.4%) who fulfilled breast-ovarian cancer testing indications had a history of other cancer types. It is well known that colorectal cancer, endometrial cancer, brain tumor, and sarcoma are associated with genetic predispositions such as MMR genes or TP53. Many high-penetrance genes also exhibit pleiotropic clinical manifestations of other common cancers. As multi-gene panel testing for all breast cancer patients was found to be cost-effective, it may be rational to expand testing to patients with cancers in the HBOC spectrum36.
Lastly, we identified double pathogenic variants in ten individuals. To date, there is limited data on double mutations in cancer patients. One study found double heterozygous variants in 1.2% of hereditary breast cancer patients37. However, the exact prevalence is still undetermined. It is unclear if patients with double mutations would have different cancer susceptibility and clinical severity compared to single mutation carriers. However, this information can be used for proper surveillance strategies for a broader spectrum of cancers in patients and at-risk family members.
With various screening methods for breast and ovarian cancer, as well as the availability of prophylactic surgeries in most regions of Thailand, this data will properly guide physicians for personalized surveillance and preventive strategies in patients or at-risk family members. Our findings will support the rationale of implementing multi-gene panel testing beyond BRCA1/2 in Thai patients with HBOC-related cancers and provide more information on the Southeast Asian population. With cost reduction and faster turnaround time, the clinical use of multi-gene panel tests for cancer will gradually increase among many low-to-middle-income countries.
This study had some limitations. Firstly, some variants were found in a small number of patients. Therefore, the clinical data associated with those variants were limited. Secondly, some clinical data was incomplete, and the follow-up duration may not have been long enough to demonstrate susceptibility to other types of cancer. Thirdly, this cohort may be biased towards breast cancer patients and away from prostate cancer. This is explained by greater awareness about genetic factors involved in breast and ovarian cancers, leading to more genetic testing in these patients. Moreover, guidelines for genetic testing in high-risk breast/ovarian cancer patients have been widely published, while genetic testing for prostate cancer has only been recommended in recent NCCN guidelines for patients with metastatic disease, strong family history, or high-risk features (e.g., very high PSA level or high Gleason score)5,38. In Thailand, genetic testing for prostate cancer is not included in current national guidelines, leading to underutilization in these patients. Finally, this was a single-center study with most patients from the central region of Thailand. However, Siriraj Hospital is a major cancer referral center that provides genetic testing services to other hospitals nationwide.
Methods
Patient recruitment and data collection
This study was approved by the Siriraj Hospital Institutional Review Board Protocol No. 418/2562(EC2) and was conducted according to Good Clinical Practice and the Declaration of Helsinki. All participants provided written informed consent. All Thai patients with any cancers in the HBOC spectrum, including breast, ovarian, pancreatic, and prostate cancers, who had germline cancer susceptibility multi-gene panel testing at Siriraj Hospital between 2016 and 2023 were included. The data of all patients with germline P/LP variants in cancer susceptibility genes were collected. Patients with known clinical syndromes of Mendelian disorders (such as neurofibromatosis, ataxia-telangiectasia, or Peutz-Jeghers syndrome) and individuals who received targeted gene testing due to known affected family members were excluded. The data regarding the types of cancers, histopathological profiles, clinical stages of cancer, age of onset, and family history were systematically collected and reviewed with 2023 NCCN guidelines for genetic/familial high-risk assessment of breast and ovarian cancers5. We calculated the positive rate of P/LP BRCA and non-BRCA variants using descriptive statistics for each cancer type. Additionally, we reported the clinical spectrum of breast cancer patients with non-BRCA P/LP variants.
Multi-gene panel testing and variant analysis
Peripheral blood specimens from each patient were collected to extract genomic DNA for sequencing (see Supplementary 4). The comprehensive cancer panel in this study included moderate-to-high penetrance genes for breast and ovarian cancers (Table 5)5,6,7,39. Sanger sequencing and Multiplex Ligation-dependent Probe Amplification (MLPA) were performed to validate the results in all identified single nucleotide variants (SNV) and copy number variants (CNV), respectively. The variant call format (VCF) and BAM files were transferred to VarSeq – VSClinical software (Golden Helix, USA) for analysis and classification. We interpreted and classified the variants following the 2015 ACMG-AMP standards and guidelines for the interpretation of sequence variants and 2020 ACMG-ClinGen technical standards for the interpretation and reporting of constitutional copy-number variants40,41. All reportable variants, including P/LP variants and VUS, were systematically verified.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author through collaboration and/or data usage agreement under the Genomics Thailand data protection and usage regulation by the Health Systems Research Institute of Thailand.
Code availability
Torrent Suite™ Software (Thermo Fisher Scientific, USA) was used for primary data analysis. SEQUENCE Pilot—SeqNext (JSI Medical Systems GmbH, Germany) was used for base calling, re-alignment, variant calling, and annotation. VarSeq—VSClinical software (Golden Helix, USA) was used for variant analysis and classification. No custom code was used to generate or process the data described in the manuscript.
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Arnold, M. et al. Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast 66, 15–23 (2022).
Nagy, R., Sweet, K. & Eng, C. Highly penetrant hereditary cancer syndromes. Oncogene. 23, 6445–70 (2004).
Garber, J. E. & Offit, K. Hereditary cancer predisposition syndromes. J. Clin. Oncol. 23, 276–292 (2005).
Daly M. B. et al. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 3.2023, NCCN clinical practice guidelines in oncology 2023 [Available from: https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf].
Shiovitz, S. & Korde, L. A. Genetics of breast cancer: a topic in evolution. Ann. Oncol. 26, 1291–1299 (2015).
Graffeo, R. et al. Moderate penetrance genes complicate genetic testing for breast cancer diagnosis: ATM, CHEK2, BARD1, and RAD51D. Breast. 65, 32–40 (2022).
Breast Cancer Association Consortium, Dorling, L. et al. Breast cancer risk genes - association analysis in more than 113,000 women. N. Engl. J. Med. 384, 428–439 (2021).
Hu, C. et al. A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med. 384, 440–451 (2021).
Kurian, A. W. et al. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer. JAMA Oncol. 4, 1066–1072 (2018).
Su, Y. et al. Characteristics of germline non-BRCA mutation status of high-risk breast cancer patients in China and correlation with high-risk factors and multigene testing suggestions. Front. Genet. 12, 674094 (2021).
Ow, S. G. W., Ong, P. Y. & Lee, S. C. Discoveries beyond BRCA1/2: multigene testing in an Asian multi-ethnic cohort suspected of hereditary breast cancer syndrome in the real world. PLoS One. 14, e0213746 (2019).
Li, J. Y. et al. Germline mutations in 40 cancer susceptibility genes among Chinese patients with high hereditary risk breast cancer. Int. J. Cancer. 144, 281–9 (2019).
Mannan, A. U. et al. Detection of high frequency of mutations in a breast and/or ovarian cancer cohort: implications of embracing a multi-gene panel in molecular diagnosis in India. J. Hum. Genet. 61, 515–22 (2016).
Singh, J. et al. Screening of over 1000 Indian patients with breast and/or ovarian cancer with a multi-gene panel: prevalence of BRCA1/2 and non-BRCA mutations. Breast Cancer Res. Treat. 170, 189–196 (2018).
Lertwilaiwittaya, P. et al. A cost-utility analysis of BRCA1 and BRCA2 testing in high-risk breast cancer patients and family members in Thailand: a cost-effective policy in resource-limited settings. Front. Public Health. 11, 1257668 (2023).
Tuffaha, H. W. et al. Cost-effectiveness analysis of germ-line BRCA testing in women with breast cancer and cascade testing in family members of mutation carriers. Genet. Med. 20, 985–994 (2018).
Lourenção, M. et al. Cost-effectiveness of BRCA 1/2 genetic test and preventive strategies: using real-world data from an upper-middle income country. Front. Oncol. 12, 951310 (2022).
Wong, E. S. Y. et al. Inherited breast cancer predisposition in Asians: multigene panel testing outcomes from Singapore. NPJ Genom. Med. 1, 15003 (2016).
Shin, H. C. et al. Detection of germline mutations in breast cancer patients with clinical features of hereditary cancer syndrome using a multi-gene panel test. Cancer Res. Treat. 52, 697–713 (2020).
Kapoor, N. S. et al. Multigene panel testing detects equal rates of pathogenic BRCA1/2 mutations and has a higher diagnostic yield compared to limited BRCA1/2 analysis alone in patients at risk for hereditary breast cancer. Ann Surg Oncol. 22, 3282–3288 (2015).
Moslemi, M. et al. The prevalence of ataxia telangiectasia mutated (ATM) variants in patients with breast cancer patients: a systematic review and meta-analysis. Cancer Cell Int. 21, 474 (2021).
Thompson, D. et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J. Natl. Cancer Inst. 97, 813–822 (2005).
Couch, F. J. et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 3, 1190–1196 (2017).
Bernstein, J. L. et al. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women’s environmental cancer and radiation epidemiology study. J. Natl. Cancer Inst. 102, 475–483 (2010).
Yadav, S. et al. Evaluation of germline genetic testing criteria in a hospital-based series of women with breast cancer. J Clin Oncol. 38, 1409–1418 (2020).
Desai, N. V., Yadav, S., Batalini, F., Couch, F. J. & Tung, N. M. Germline genetic testing in breast cancer: rationale for the testing of all women diagnosed by the age of 60 years and for risk-based testing of those older than 60 years. Cancer. 127, 828–833 (2021).
Manahan, E. R. et al. Consensus guidelines on genetic‘ testing for hereditary breast cancer from the american society of breast surgeons. Ann. Surg. Oncol. 26, 3025–3031 (2019).
Whitworth, P. W. et al. Clinical utility of universal germline genetic testing for patients with breast cancer. JAMA Netw. Open. 5, e2232787 (2022).
Culver, J. O. et al. Integration of universal germline genetic testing for all new breast cancer patients. Ann. Surg. Oncol. 30, 1017–1025 (2023).
LaDuca, H. et al. A clinical guide to hereditary cancer panel testing: evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet. Med. 22, 407–415 (2020).
Eoh, K. J. et al. Detection of germline mutations in patients with epithelial ovarian cancer using multi-gene panels: beyond BRCA1/2. Cancer Res Treat. 50, 917–925 (2018).
Lhotova, K. et al. multigene panel germline testing of 1333 Czech patients with ovarian cancer. Cancers. 12, 956 (2020).
Bonadona, V. et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 305, 2304–2310 (2011).
Engel, C. et al. Risks of less common cancers in proven mutation carriers with lynch syndrome. J Clin Oncol. 30, 4409–4415 (2012).
Sun, L. et al. A cost-effectiveness analysis of multigene testing for all patients with breast cancer. JAMA Oncol. 5, 1718–1730 (2019).
Megid, T. B. C., Barros-Filho, M. C., Pisani, J. P. & Achatz, M. I. Double heterozygous pathogenic variants prevalence in a cohort of patients with hereditary breast cancer. Front. Oncol. 12, 873395 (2022).
Tuffaha H. et al. Guidelines for genetic testing in prostate cancer: a scoping review. Prostate Cancer Prostatic Dis. https://doi.org/10.1038/s41391-023-00676-0 (2023).
Lertwilaiwittaya, P. et al. Thai patients who fulfilled NCCN criteria for breast/ovarian cancer genetic assessment demonstrated high prevalence of germline mutations in cancer susceptibility genes: implication to Asian population testing. Breast Cancer Res. Treat. 188, 237–248 (2021).
Riggs, E. R. et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 22, 245–57 (2020).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Acknowledgements
We thank all participants for their cooperation and contribution to our study. We thank all physicians and health professionals for their patient’s clinical care. We also thank the Genomics Thailand Cancer Program and Research University Network (RUN)—Thailand for the administrative support. This work was supported by Health Systems Research Institute—Genomics Thailand Initiative Grant to MP, CLi; Siriraj Core Research Facility (SiCRF) and Strategic Project Grant to MP; Siriraj Chalermphrakiat Grant to WTh, CLi, and MP; Thanapat Fund (D003752) to MP. The funders had no role in study design, data collection and analysis, publication decisions, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
MP, PL, CLi, ER, and CK contributed to the conception and design of the study. PN, PD, CM, WTa, KPo, SW, WTi, NP, CLe, JW, JS, KPu, and ER performed the experiments. PN, PD, PL, PM, CM, WTa, KPo, SW, WTi, NP, CLe, JW, JS, JK, KPu, CK, WTh, ER, CLi, and MP collected and analyzed the data. CK and MP wrote and substantially revised the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kansuttiviwat, C., Lertwilaiwittaya, P., Roothumnong, E. et al. Germline mutations of 4567 patients with hereditary breast-ovarian cancer spectrum in Thailand. npj Genom. Med. 9, 9 (2024). https://doi.org/10.1038/s41525-024-00400-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-024-00400-4