Abstract
Adiponectin, encoded by ADIPOQ, is an insulin-sensitizing, anti-inflammatory, and renoprotective adipokine that activates receptors with intrinsic ceramidase activity. We identified a family harboring a 10-nucleotide deletion mutation in ADIPOQ that cosegregates with diabetes and end-stage renal disease. This mutation introduces a frameshift in exon 3, resulting in a premature termination codon that disrupts translation of adiponectin’s globular domain. Subjects with the mutation had dramatically reduced circulating adiponectin and increased long-chain ceramides levels. Functional studies suggest that the mutated protein acts as a dominant negative through its interaction with non-mutated adiponectin, decreasing circulating adiponectin levels, and correlating with metabolic disease.
Similar content being viewed by others
Introduction
Adiponectin is an adipocyte-derived hormone with pleiotropic actions that promote insulin sensitivity, inhibit cell death, and decrease inflammation1. Adiponectin forms an obligate trimer and circulates as trimers, hexamers, and high-molecular weight multimers that target various tissues and cell types, including liver, kidney, cardiac myocytes, and pancreatic β cells. Levels of adiponectin are decreased in obesity and may contribute to a chronic state of inflammation that leads to insulin resistance, type 2 diabetes, coronary artery disease, myocardial infarction, nonalcoholic steatohepatitis, and kidney disease2,3,4. Recently, the antiapoptotic, insulin-sensitizing, glucose lowering, anti-inflammatory, and anti-steatotic effects of adiponectin have been linked to its role in sphingolipid metabolism and its receptor-mediated activation of ceramidase activity, which reduce levels of lipotoxic ceramides5,6.
Previous studies have identified common variants in or near ADIPOQ, the gene that encodes adiponectin, that are associated with aberrant adiponectin levels7, obesity8, type 2 diabetes7,9, and diabetic kidney disease10,11. Rare ADIPOQ amino acid substitution mutations have also been reported in individuals with diabetes and hypoadiponectinemia12,13. Here, we describe the first multigenerational family with a protein-truncating mutation in ADIPOQ (p.Gly93GlufsTer73), diabetes, and end-stage renal disease. Carriers of this mutation have dramatically reduced circulating adiponectin and increased long-chain ceramide levels. Functional studies show that this mutation acts via a dominant negative mechanism, with the wild-type and mutant proteins interacting, leading to decreased levels of cellular and secreted adiponectin. Our findings support adiponectin’s protective role and suggest that its genetic loss leads to ceramide accumulation which, in turn, contributes to diabetes and progression of renal disease.
Results
Identification of ADIPOQ as a candidate gene for diabetes and end-stage renal disease
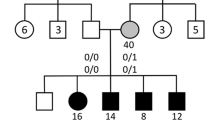
The goal of this study was to identify a potential genetic cause of disease in a mutigenerational family with diabetes and prominent end-stage renal disease. Included among the cases in this family were three affected sibling pairs, two affected parent-offspring pairs, two affected first-cousin pairs, and two pairs of affected first-cousins-once-removed (Fig. 1a and Table 1). Using unified linkage analysis and rare variant association testing, where the enrichment of rare variants among these cases was compared to 524 ethnically matched background controls, we identified a heterozygous 10-nucleotide deletion in exon 3 of ADIPOQ (chromosome 3: 186,572,030; CCCGAGGCTTT→C) shared by all six affected family members (Fig. 1a–d, Supplementary Figs. 1–3, and Supplementary Tables 1–4). This rare deletion (p.Gly93GlufsTer73) was seen only once among 56,810 exome and genome sequences from non-Finnish Europeans reported in the Genome Aggregation Database14 and creates a frameshift that results in premature truncation of the adiponectin protein, generating a novel peptide that includes 73 abnormally coded terminal amino acids (Fig. 1b and Supplementary Fig. 4).
a The family pedigree, status of diabetes (shaded) and end-stage renal disease (ESRD; red outline), and the ADIPOQ mutation (p.Gly93GlufsTer73). Carriers of the wild-type ADIPOQ insertion (I) or mutated ADIPOQ deletion (D; red) are indicated. b The structure of the wild-type (top) and mutant (bottom) adiponectin protein. Wild-type adiponectin monomers consist of a 244 amino acid protein composed of four domains; an N-terminal signal sequence (19 amino acids), a variable region (23 amino acids), a collagenous domain (66 amino acids), and a C-terminal globular domain (136 amino acids). The 10-nucleotide deletion (CCCGAGGCTTT→C, indicated as ∆CCGAGGCTTT) at amino acid 93 creates a frameshift that truncates the adiponectin protein and generates a novel peptide that terminates 73 amino acids after this deletion. c The PHEVOR plot using Human Phenotype Ontology (HPO) terms and connections to Gene Ontology terms to prioritize potentially damaging alleles, using terms for a) kidney disease: HP:0000077 (Abnormality of the kidney), HP:0000112 (Nephropathy), and HP:0003774 (Stage 5 chronic kidney disease), and b) diabetes: HP:0000819 (Diabetes) and HP:0005978 (Type 2 diabetes mellitus) and a combination of the pVAAST p values and PHEVOR scores. PHEVOR scores (y-axis) for each gene (dot) are plotted across the genome (x-axis, chromosomes 1-Y). d The chromatogram from Sanger sequencing of a non-carrier from the family (WT) and the 6 carriers identified through whole-genome sequencing and pVAAST/PHEVOR analysis.
In addition to the six affected family members with diabetes and end-stage renal disease, eight additional family members were available for this study. Sanger sequencing confirmed the carrier status of the six affected family members and identified four additional carriers and four non-carriers of the ADIPOQ mutation in the family (Fig. 1a and d). Five of these carriers (Family Members III:3, III:8, IV:2, V:1, and V:2) had a diagnosis of diabetes either shortly before enrollment to the Joslin Study on the Genetics of Type 2 Diabetes15 or following their participation in this study; neither, likely due to their short duration of diabetes at enrollment and the lack of follow-up, developed diabetic kidney disease or end-stage renal disease to our knowledge. The two other carriers (Family Members III:2 and IV:1) were nondiabetic at enrollment and no follow-up data are available. Among carriers of the ADIPOQ mutation, only three possessed either one human leukocyte antigen (HLA) DR3 or DR4 risk allele, suggesting that their diabetes is likely not due to the autoimmune form of this disease (Table 1).
Circulating adiponectin and ceramide levels of mutant ADIPOQ carriers and noncarriers
To investigate whether the ADIPOQ mutation alters the level of circulating adiponectin, we measured adiponectin in serum from all 14 family members. Carriers of this truncating mutation had significantly reduced circulating adiponectin, less than 20% of the levels found in noncarriers (Fig. 2a). We also fractionated serum using fast protein liquid chromatography (FPLC)-based gel filtration and each fraction was probed for adiponectin by Western blot. Carriers of the mutation lacked high molecular weight (HMW) adiponectin complexes (Fig. 2b and Supplementary Fig. 5a, b), while HMW adiponectin was the most abundant isoform in noncarriers. As adiponectin’s role as a protective adipokine has been attributed to its ability to reduce lipotoxic ceramides5, we used liquid chromatography with tandem mass spectrometry (LC-MS/MS)16 to analyze ceramide level in carriers and noncarriers from this family. As anticipated, we observed that carriers of the ADIPOQ mutation had on average a 35% increase in C16.0 ceramide levels compared to noncarriers (Fig. 2c) and even greater increases relative to nondiabetic controls (Supplementary Fig. 6).
a The level of circulating adiponectin, measured by enzyme-linked immunosorbent assay (ELISA), in noncarriers (black, n = 7) and carriers (red, n = 8) of the ADIPOQ mutation (two-tailed t-test, α = 0.05, *p value ≤ 0.05, error bars SEM). b The intensity values of bands from western blotting for noncarrier (black, n = 2) and carrier (red, n = 2) patients. Carriers of the ADIPOQ mutation lack bands in FPLC fractions associated with high molecular weight (HMW) adiponectin complexes (error bars SEM). Representative western blots are shown and area under the curve was calculated for each peak, HMW, low molecular weight (LMW), and adiponectin trimers (2-way ANOVA with Fisher’s LSD correction, α = 0.05, p = 0.018, error bars are SEM). c The levels of circulating adiponectin, measured by ELISA, in noncarriers (black, n = 4) and carriers (red, n = 10) of the ADIPOQ mutation relative to C16.0 ceramide levels, measured by LC-MS/MS. d Western blot showing wild-type and mutant adiponectin produced in HEK293T cells. The upper blot represents cell lysates and the lower blot shows the two proteins excreted into the cell media. The lanes denoted with a+ were samples taken from cells treated with 20 µg/mL of MG132. Beta actin was used as a loading control. e The FRET efficiency calculated from acceptor bleaching and calculating the increase of intensity of the donor fluorophore. Clover-ADIPOQ and mRuby2-ADIPOQ serve as a positive control for protein-protein binding and FRET. The Clover-ADIPOQ and mutant mRuby2-ADIPOQ cells show no significant FRET efficiency, though very little mutant mRuby2-ADIPOQ is observed in the cells. This is ameliorated by the addition of MG132 (20 µg/mL) blocking mutant mRuby2-ADIPOQ from being degraded. The number of cells counted for quantification is denoted above each bar in the graph. (Asterisks represent significance by a one-sample t test indicating that the chance of FRET occurring is above zero, α = 0.05, *p value ≤ 0.05, **p value ≤ 0.01, ****p value ≤ 0.0001. The large star represents a significant difference between non-treated and MG132 treated Clover-ADIPOQ and mutant mRuby2- ADIPOQ cells by two-tailed t test, α = 0.05, p = 0.04, error bars SEM). The panel also shows an intensity modulated ratiometric image of cells treated with MG132 that produce both Clover-ADIPOQ and the mutant mRuby2-ADIPOQ proteins. Red pseudo color shows pixels in which the donor intensity increases after photobleaching, indicating a FRET signal as the proteins interact.
Functional characterization of the ADIPOQ mutation
Previous studies have shown that missense point mutations in adiponectin impair multimerization, which is critical for formation of functional trimers, hexamers, and high molecular weight multimers12,13. As the ADIPOQ mutation significantly alters adiponectin structure, we hypothesized that this mutation likely disrupts adiponectin function. To investigate this, we developed gene expression constructs to allow tight expression of wild-type human adiponectin and the ADIPOQ mutant form of the protein within a single vector in human stable transfected embryonic kidney 293T (HEK293T) cells (Supplementary Fig. 7). Expression of mutant adiponectin can only be visualized when protein degradation is inhibited by MG132 (Fig. 2d and Supplementary Fig. 5c). Cells that express both the wild-type and the mutant protein express less adiponectin inside the cell, with reduced amounts secreted into the media. In the presence of MG132, the mutant protein can be visualized and cells producing both wild-type and mutant protein show a significant reduction of adiponectin within the cell and excreted to the media. To determine whether mutant and wild-type adiponectin interact, we conducted FRET assays using adiponectin tagged with either Clover or mRuby2 (Supplementary Fig. 8). As expected, acceptor-bleaching of living cells using confocal microscopy revealed that these adiponectin monomers interacted, forming multimers (Fig. 2e). When the mRuby2-tag was added to the mutant adiponectin construct, rather than the non-mutated isoform, FRET efficiency decreased substantially, likely due to degradation of the mutant form of the protein by the cells. The proteasome inhibitor MG132 increased FRET efficiency in these cells. The intensity modulated ratiometric image (Fig. 2e) represents MG132-treated cells that express both wild-type and mutant adiponectin. The sensitized emission model of FRET showed moderate levels of heterologous protein interaction that increased substantially following treatment with MG132 (Supplementary Fig. 8). Taken together, these data show that adiponectin and the truncated variant produced by the ADIPOQ mutation interact, leading to decreased stability of the wild-type adiponectin. It is in this way that the mutant adiponectin exerts a dominant-negative effect that results in significantly reduced adiponectin levels.
Discussion
We report the identification of a rare truncating mutation in ADIPOQ in a multigenerational family enriched for diabetes and end-stage renal disease. Although we cannot rule out that other renal pathologies account for the kidney disease observed in this family, as no other monogenic kidney disease gene (e.g., PKD1, PKD2, COL4A3/4/5, etc.) emerged from our genetic analysis of this family, suggesting that other heritable forms of kidney disease are unlikely, diabetes is the suspected cause. Our data show that this mutation results in dramatically reduced circulating adiponectin (<20% of normal levels) and high ceramide levels (more than a 30% increase in C16.0 ceramides compared to noncarriers). The ADIPOQ mutation, located in exon 3 (i.e., the terminal exon) of the encoded protein, causes a frameshift in the sequence encoding the collagenous domain of adiponectin and disrupts formation of its globular domain. Given its location, we hypothesize that the resulting mutant protein escapes the usual mechanism of nonsense-mediated decay and is able to form characteristic multimers, but targets them for degradation. The resultant reduction in functional adiponectin multimers leads to increased ceramide levels. The low-adiponectin, high ceramide state in this family is associated with diabetes and progressive loss of kidney function.
Rare missense mutations, primarily at conserved amino acid residues in adiponectin’s collagenous and globular domains were previously identified in individuals with type 2 diabetes and hypoadiponectinemia12,13,17,18,19. Interestingly, similar to carriers of the ADIPOQ mutation identified in our study, several point mutations, within the globular domain of adiponectin, cause low circulating levels of adiponectin18. Most notably, adiponectin p.R112C and adiponectin p.I164T show 27% and 32% of the circulating adiponectin seen in wild-type adiponectin carriers, respectively. In this study, ADIPOQ mutation carriers show 24% of the circulating adiponectin of unaffected family members. Each mutant adiponectin similarly disturbs multimer assembly and secretion of the wild-type protein. Unlike the truncated adiponectin studied here, which is nearly undetectable in the absence of MG132, these previously identified point mutants form stable monomers with levels of intracellular protein expression similar to wild-type adiponectin. A portion of adiponectin is known to remain within the adipocyte, and can maintain potent ceramide-lowering biological activity.
Expanding on these findings, in addition to reporting the first strongly pathogenic mutation in ADIPOQ that leads to familial hypoadiponectinemia, we also demonstrate that this mutation results in a prominent increase in C16.0 ceramide levels. Although it remains unclear why C16 ceramides are more pathological than other ceramide species, recent studies have demonstrated that the Cers6 enzyme that generates C16 ceramides is selectively upregulated in obese humans and mice20 and these ceramides uniquely trigger mitochondrial dysfunction21. Conversely, we have linked adiponectin and adiponectin receptors AdipoR1 and AdipoR2 to ceramide catabolism and C16 ceramides are uniquely altered by overexpression of adiponectin, AdipoR1, or AdipoR2 in preclinical studies22. Thus, higher C16 ceramides associated with the ADIPOQ mutation are likely a result of diminished ceramidase activity and ceramide catabolism resulting from the inability of reduced functional adiponectin multimers to stimulate its two receptors, ADIPOR1 and ADIPOR25. In the total absence of adiponectin, these receptors fail to catalyze ceramide degradation6. Importantly, ceramides, a class of deleterious sphingolipids, have previously been shown to contribute to cellular dysfunction that causes impaired insulin action, induces β cell death, and exacerbates kidney failure23.
Several recent studies have demonstrated associations between abnormal long-chain ceramides, as well as other sphingolipids species in this pathway, and albuminuria, estimated glomerular filtration rate decline, and end-stage renal disease in subjects with diabetes24,25,26. Further supporting the role of ceramide biosynthesis in diabetic kidney disease, large-scale genetic studies have linked a common deleterious missense variant in ceramide synthase-2 (CERS2), a ceramide-synthesizing gene in the sphingolipid biosynthesis pathway, with reduced renal function and diabetic kidney disease risk27,28. While the heritability of ceramide levels has been estimated to be as high as 0.52, our understanding of the genetic factors that underlie variation of ceramide levels is woefully incomplete29. Indeed, variability in ceramide levels, even among carriers of the ADIPOQ mutation, suggest that this is likely a polygenic trait influenced by a network of genetic, as well as environmental, factors. Of note, patient III:2, successfully avoided diabetes and is the only carrier of normal body weight. Conversely, many noncarriers in this study were also diabetic. Maternal-fetal environment30, shared dietary patterns amongst family members, and epigenetic alterations31 could contribute to some degree of diabetes risk. Notably, adiponectin null mice even produce breastmilk that is proinflammatory, which can alter the health status of their offspring32. Although ceramides have clearly been linked to diabetic kidney disease, the underlying factors that lead to ceramide biology dysfunction are largely unknown, highlighting a need for additional research in this area.
In summary, we identified the first multigenerational family with a protein truncating mutation in ADIPOQ, diabetes, and end-stage renal disease. This mutation impairs adiponectin multimer formation, dampens intracellular protein expression, and impacts adiponectin’s role in glucose and lipid metabolism. Our findings further implicate adiponectin dysfunction in diabetes and diabetic kidney disease and link abnormalites in circulating adiponectin with elevated ceramide levels through impaired activation of its cognate receptors. Importantly, compounds exist (e.g., AdipoRon33) that activate adiponectin receptors and lower ceramide abundance. For this family, adiponectin mimetics represent a potential personalized medicine approach to counteract deficient adiponectin levels and increased ceramide levels due to this mutation. More broadly, interventions to lower ceramides could prove to be novel therapies for treating diabetes or slowing the progression of diabetic kidney disease.
Methods
Family enriched for diabetes and end-stage renal disease
Individuals from the family examined in this study were enrolled in the Joslin Study on the Genetics of Type 2 Diabetes between 1993 and 200315. Briefly, between 1993 and 2003, families with an apparent autosomal dominant mode of inheritance of type 2 diabetes (T2D), irrespective of their nephropathy status, were recruited to the Joslin Study of Genetics of Nephropathy in Type 2 Diabetes Family Collection through T2D probands receiving medical care at the Joslin Clinic. The protocols and written informed consent procedures used in this study were approved by the Committee on Human Subjects of the Joslin Diabetes Center.
All participating family members provided written informed consent prior to participating in this study. All family members were of European ancestry. After obtaining informed written consent, trained recruiters administered previously described study protocols that included a structured interview, seated blood pressure measurements, and the collections of blood and urine samples. All blood and urine specimens have been stored at −80 °C since the time of their collection. ESRD status for members of this family was updated through the United States Renal Data System.
Genetic analyses
Whole genome sequencing of DNA samples from six individuals with ESRD was performed on the Illumina HiSeq2500 platform available at Macrogen (Rockville, MD). FASTQ files from paired-end sequencing were subsequently aligned to the Hg19/GRCh37 reference genome using the Sentieon Genomics DNASeq software pipeline per the developer’s guidelines34. The DNASeq pipeline incorporates the Broad Institute’s BWA-GATK Best Practices Workflow but performs alignments and variant calling in a computationally efficient process. The six individual samples were jointly genotyped with 524 ethnically matched background controls comprised of 291 individuals from the 1000 Genomes Project and 233 healthy samples from the Utah Genome Project35. The pipeline created individual genomic variant call files and a final jointly-called variant call file (VCF). Sample qualities in final VCF were evaluated using Peddy36 to determine and confirm sex, relatedness, heterozygosity and ancestry of each individual to identify any potential sample quality issues. The VCF was functionally annotated using ANNOVAR for downstream analyses37.
A unified linkage analysis and rare variant association testing using pedigree Variant Annotation, Analysis, and Search Tool (pVAAST), version 2.2.0, was performed on six individual sample target genomes and 524 ethnically matched background controls35. pVAAST incorporates pedigree information into disease-gene prioritization procedures that incorporates amino acid substitution and calculates burden of each gene candidate. pVAAST was performed using 1 × 10−7 permutations and configured to evaluate penetrance values in the range of 0.5–0.995, with LOD and CLRT filters removed. In keeping with best practices from the developers of this tool, we considered any gene with a variant achieving a pVAAST p value < 0.005 as a potential positive signal. Additional reranking of pVAAST outputs occurred using Phenotype Driven Variant Ontological Re-ranking tool (PHEVOR) using Human Phenotype Ontology (HPO) terms and connections to Gene Ontology (GO) terms to prioritize potentially damaging alleles, using the HPO terms a) for kidney disease: HP:0000077 (Abnormality of the kidney), HP:0000112 (Nephropathy), and HP:0003774 (Stage 5 chronic kidney disease), and b) for diabetes: HP:0000819 (Diabetes) and HP:0005978 (Type 2 diabetes mellitus) and a combination of the pVAAST p value and Phevor score38.
Sanger sequencing confirmation of the ADIPOQ variant was performed in all 6 individuals with ESRD in this family as well as 8 additional family members for whom DNA was available. The genomic region covering the variant was PCR amplified using the Qiagen Taq PCR amplification kit (Qiagen, Valencia, CA) per the manufacturer’s protocol and the following primer pair: forward 5′–GGCTGTAACCAACCTAGGCAGG–3′ and reverse 5′–AGGCAAAGTAGTACAGCCCAGG–3′. Sanger sequencing was performed using BigDye Terminator v3.1 Cycle Sequencing chemistry (Life Technologies, Carlsbad, CA). Capillary electrophoresis was performed by the DNA Sequencing Core Facility at the Health Science Center at the University of Utah using a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). The resulting chromatograms were analyzed using Sequencher version 5.4.6 (Gene Codes Corporation, Ann Arbor, MI).
All 14 family members were genotyped on the HumanCore BeadChip (Illumina, San Diego, CA, USA), which contains 250,000 genome-wide tag SNPs (and other variants) and over 200,000 exome-focused variants, by the DNA Sequencing Core Facility at the Health Science Center at the University of Utah. All samples were passed through a stringent quality control protocol that included filtering for low-quality variants (e.g., call rates <95% and excessive deviation from Hardy–Weinberg equilibrium).
HLA alleles DR3 (haplotype DQA1*05:01-DQB1*02:01) and DR4 (haplotype DQA1*03-DQB1*03:02) were predicted for all 14 family members using genome-wide genotyping data, the SNP2HLA software39, and genome-wide genotyping data from the HLA-region and the Type 1 Diabetes Genome Consortium’s reference genotype-HLA-allele panel.
Circulating adiponectin
Circulating adiponectin levels were measured in serum from 14 family members (including 4 noncarriers and 10 carriers of the ADIPOQ mutation) using an enzyme-linked immunosorbent assay (ELISA) (Millipore EZHADP-61K). Protein concentration was assessed using a BCA kit (Pierce) prior to load equivalent protein levels from all patients. The ELISA was measured using a ThermoFisher Varioskan Lux Microplate Reader.
Fast protein liquid chromatography (FPLC)
FPLC was performed as previously described40. Serum (50 µL) from two representative noncarrier or carrier siblings was injected into an ÄKTA Go FPLC (GE Healthcare). A Superdex 200 10/300 GL column (GE Healthcare) was used to separate adiponectin complexes in HEPES/Ca2+ buffer (25 mM HEPES; 150 mM NaCl; and 1 mM CaCl2, pH 7.4). 250 µL fractions were collected over a 20 mL retention volume. The retention volumes found to contain adiponectin were then used to run Western blots to determine HMW, LMW and trimeric adiponectin. Samples were run on an SDS-Page gel (BioRad Criterion TGX) after being reduced in Laemmli and 355 mM 2-mercaptoethanol and boiled for 10 min. The gel was transferred to PVDF membrane (BioRad). The membranes for each patient were blocked in 5% BSA then probed for adiponectin overnight with rabbit polyclonal anti-adiponectin at a 1:1000 dilution (Abcam, ab75989). There were washed then stained with goat-anti-rabbit AlexaFluor Plus 680 (Invitrogen) at a 1:4000 dilution. Then washed again and imaged on a ThermoFisher iBright system. Western blots derive from the same experiment and were processed in parallel.
Lipidomic analysis
Ceramide levels were measured using targeted lipidomic profiling and liquid chromatography with tandem mass spectrometry (LC-MS/MS) at the University of Utah’s Metabolomics Core. Analysis of lipid species, including C16.0 ceramides, was performed as previously been described16. Briefly, these determinations were obtained from serum specimens from 14 family members as well as 25 unrelated nondiabetic controls using targeted LC-MS/MS platforms at the University of Utah’s Metabolomics Core with each lipid normalized to an internal standard using an established and previously described method developed by the Summers lab16. Statistical analysis was performed using Student’s t test in SAS for Windows, version 9.2 (SAS Institute, Cary, NC) following log10 transformation of these data and results are considered significant at the P < 0.05 level.
Gene expression vectors
Gene sequences for human wild-type and mutant ADIPOQ were cloned into the pVitro2 vector (InvivoGen) expressing blasticidin resistance (Fig. S6). The vector is designed for dual gene expression with both genes driven by two human ferritin composite promoters. Following the addition of wild-type ADIPOQ is an IRES followed by the bsr gene allowing for blasticidin resistance. For cells that express only wild-type or mutant ADIPOQ, the gene sequences were disrupted with a premature stop codon so that only one variant of adiponectin is produced. The vectors were transfected into HEK293T cells (ATCC, CRL-11268) using JetPrime (Genesee Scientific). Twenty-four hours after transfection, 20 µg/mL blasticiden was added to the cell culture media. The cells were grown and split three times to ensure stable expression of the genes. The cells were maintained in 10 cm cell culture dishes in DMEM with 10% FBS, pen/strep, and blasticidin. For cells treated with the proteasome inhibitor MG132 (Sigma), healthy cells were treated for 12–18 h with a concentration of 20 µM in FBS free media.
Western blotting
Stable expressing cells were grown to 80–90% confluency. Then media was removed and supplemented with DMEM with no FBS. Twelve to eighteen hours later the media was collected with the addition of protease inhibitors (Roche) and put-on ice. The cells were lysed using 200 µL of ice cold radioimmunoprecipitation assay buffer containing protease inhibitors (Roche). The cells were vortexed and lightly sonicated to form a homogeneous solution. Protein concentration was assessed using a BCA kit (Pierce). For traditional Western blotting, 2 µg of protein was reduced using Bolt Reducing Agents (ThermoFisher) and heated for 5 min. Protein was run on Invitrogen Bolt 4–12% Bis-Tris gels (ThermoFisher) and then transferred to a nitrocellulose membrane using the Invitrogen Power Blotter. The membranes were then processed for immunodetection using the iBind Western System. Primary antibodies were selected to target adiponectin (Rabbit polyclonal anti-adiponectin from Abcam, ab75989) and beta-actin as a loading control (Mouse monoclonal anti-beta-actin from Cell Signaling, 3700S). Both were used at a 1:1000 dilution. The adiponectin antibody selected for use was made using a peptide that falls within both the wild-type and mutant adiponectin protein. Antibodies targeting the N-terminus and the C-terminus of adiponectin were also used to confirm that the mutant protein is not detected when using a C-terminus antibody (data not shown). The secondary antibodies used were Alexa-fluor Plus 488 Anti-Mouse (ThermoFisher, A32723) and Alexa-fluor Plus 800 Anti-Rabbit (ThermoFisher, A32735) at a 1:2000 dilution. The blots were imaged on a ThermoFisher iBright system. All blots derive from the same experiment and were processed in parallel.
FRET constructs and analysis
To assess if wild-type adiponectin and the mutant adiponectin protein could interact, we designed constructs with each protein tagged with a FRET sensor (Fig. S7). We chose a green fluorescent protein derivative called Clover and a red fluorescent protein derivative called mRuby2 to act as our donor/acceptor pair. Both Clover and mRuby2 were designed and proven to be excellent FRET sensors41. Either Clover or mRuby2 were attached to the protein via a linker sequence at the N-termina (since the same C-terminus is not present in the mutant protein). Individual wild-type adiponectin or mutant adiponectin were labeled with Clover or mRuby2 as single expressing constructs to gather spectral data from individual fluorophores. A double expressing construct with wild-type adiponectin labeled with Clover and wild-type adiponectin labeled with mRuby served as a FRET positive control since it is known that adiponectin forms multimers with itself. A double expressing construct with wild-type adiponectin labeled with Clover and mutant adiponectin labeled with mRuby2 served as the experimental group.
FRET readings were recorded via two methods, acceptor bleaching and sensitized emission.
Acceptor bleaching model
In this model, the acceptor fluorophore is photobleached and donor fluorophore intensities are taken prior and after the photobleaching has occurred. If FRET is occurring, the energy that would have gone toward exciting the acceptor remains with the donor, meaning that the donor intensity is increased after photobleaching the acceptor. The stable expressing cells were grown in coverslip bottom chamber slides using FluoroBrite DMEM (Gibson) with 2% FBS. The slides were mounted into a Pathology Devices, Inc stage heater kept at 37 °C and imaged on a Nikon A1R confocal microscope with a 40× oil immersion objective. Cells were quickly identified and focused using epifluorescence. A pre-bleach image was obtained using a 488 nm argon gas laser to excite Clover and a 561 nm sapphire diode laser to excite mRuby2 using predetermined laser settings for adequate imaging. After a pre-bleach image was obtained the 488 nm laser was turned off and the 561 nm laser was set to 100% power and allowed to excite the cells for 10 min to bleach the mRuby2 signal. Afterwards, a post-bleach image was acquired using the same laser and acquisition settings as the pre-bleach image. Cells containing the empty vector that produce neither Clover nor mRuby2 were imaged to provide background intensities of the cells. The intensity for Clover and mRuby2 in each cell was measured in FIJI imaging software in both the pre- and post-bleach images. The average background intensity from non-expressing cells was subtracted from the intensity measurements of the pre- and post-bleach images. Only cells that had a ≥10% reduction in mRuby2 intensity were used for quantification. FRET efficiency was calculated by \(1 - \frac{{F_{da}}}{{F_d}}\), where Fda is the intensity of the donor fluorophore before bleaching and Fd is the intensity of the donor fluorophore after the 10-min bleach. The FRET efficiency percent was graphed using Prism. Intensity modulated ratiometric images were created using RatioImage in MatLab.
Sensitized emission model
In this model, the intensity of the acceptor fluorophore is measured to determine if it gains intensity when the donor fluorophore is physically close. The stable expressing cells were grown in a Greiner µClear bottom black-walled 96-well plate using DMEM with 10% FBS, pen/strep, and blasticidin ensuring that each FRET construct (individual expressing cells and dual Clover/mRuby2 expressing cells) were in triplicate. Once the cells reached 70–80% confluency, the media was replaced with FluoroBrite DMEM (Gibson) with no FBS. The cells were treated with MG132 at this time and incubated for 12–18 h. The plate was then analyzed using a ThermoFisher Varioskan Lux Microplate Reader with the chamber heated to 37 °C and maintaining 5% CO2. The Varioskan was set to read 29 readings from the bottom of each well. To detect Clover, intensity excitation was set to 505 nm and emission detection was at 515 nm. To detect mRuby2, intensity excitation was set to 559 nm and emission detection was at 600 nm. FRET readings for dual Clover/mRuby2 expressing cells were taken with excitation set at 505 nm, to excite Clover and emission detection was set at 600 nm to read mRuby2 fluorescence. Spectral data with each excitation setting were also obtained to verify that the fluorophores emitted at their ideal peak emission curve (data not shown). Fluorescent readings were background subtracted from readings from cells with the empty vector. This FRET method requires subtracting out the spectral bleed-through derived from the donor emission spectrum overlap with the acceptor and the direct excitation of the acceptor by the donor’s excitation light to verify actual FRET. The corrected FRET ratio (FC) was obtained by:
where DSBT is the donor spectral bleed-through and the value obtained from the wild-type adiponectin tagged with Clover in the 505–600 nm reading. ASBT is the acceptor spectral bleed-through and is obtained from the mutant adiponectin tagged with mRuby2 in the 505–600 nm reading. Acceptor intensity is the measure from the 559 nm excitation and 600 nm emission reading. The corrected FRET ratio was graphed using Prism. The experiment was repeated two other times with similar results.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. All sequencing data from this study are publicly available from dbGaP (accession phs002959.v1).
References
Wang, Z. V. & Scherer, P. E. Adiponectin, the past two decades. J. Mol. Cell Biol. 8, 93–100 (2016).
Colca, J. R. & Scherer, P. E. The metabolic syndrome, thiazolidinediones, and implications for intersection of chronic and inflammatory disease. Mol. Metab. 55, 101409 (2022).
Straub, L. G. & Scherer, P. E. Metabolic messengers: adiponectin. Nat. Metab. 1, 334–339 (2019).
Sweiss, N. & Sharma, K. Adiponectin effects on the kidney. Best. Pract. Res. Clin. Endocrinol. Metab. 28, 71–79 (2014).
Holland, W. L. et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat. Med. 17, 55–63 (2011).
Holland, W. L. et al. Inducible overexpression of adiponectin receptors highlight the roles of adiponectin-induced ceramidase signaling in lipid and glucose homeostasis. Mol. Metab. 6, 267–275 (2017).
Hivert, M. F. et al. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes 57, 3353–3359 (2008).
Stumvoll, M. et al. Association of the T-G polymorphism in adiponectin (exon 2) with obesity and insulin sensitivity: interaction with family history of type 2 diabetes. Diabetes 51, 37–41 (2002).
Vionnet, N. et al. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am. J. Hum. Genet. 67, 1470–1480 (2000).
Moczulski, D. K., Rogus, J. J., Antonellis, A., Warram, J. H. & Krolewski, A. S. Major susceptibility locus for nephropathy in type 1 diabetes on chromosome 3q: results of novel discordant sib-pair analysis. Diabetes 47, 1164–1169 (1998).
He, B. et al. Association of genetic variants at 3q22 with nephropathy in patients with type 1 diabetes mellitus. Am. J. Hum. Genet. 84, 5–13 (2009).
Waki, H. et al. Impaired multimerization of human adiponectin mutants associated with diabetes. Molecular structure and multimer formation of adiponectin. J. Biol. Chem. 278, 40352–40363 (2003).
Jungtrakoon, P. et al. Novel adiponectin variants identified in type 2 diabetic patients reveal multimerization and secretion defects. PLoS One 6, e26792 (2011).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Fogarty, D. G., Rich, S. S., Hanna, L., Warram, J. H. & Krolewski, A. S. Urinary albumin excretion in families with type 2 diabetes is heritable and genetically correlated to blood pressure. Kidney Int. 57, 250–257 (2000).
Poss, A. M. et al. Machine learning reveals serum sphingolipids as cholesterol-independent biomarkers of coronary artery disease. J. Clin. Investig. 130, 1363–1376 (2020).
Hara, K. et al. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes 51, 536–540 (2002).
Kondo, H. et al. Association of adiponectin mutation with type 2 diabetes: a candidate gene for the insulin resistance syndrome. Diabetes 51, 2325–2328 (2002).
Vasseur, F. et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum. Mol. Genet. 11, 2607–2614 (2002).
Turpin, S. M. et al. Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell Metab. 20, 678–686 (2014).
Hammerschmidt, P. et al. CerS6-derived sphingolipids interact with mff and promote mitochondrial fragmentation in obesity. Cell 177, 1536–1552 e23 (2019).
Ye, R. et al. Adiponectin is essential for lipid homeostasis and survival under insulin deficiency and promotes beta-cell regeneration. Elife 3, e03851 (2014).
Summers, S. A. Could ceramides become the new cholesterol? Cell Metab. 27, 276–280 (2018).
Tofte, N. et al. Lipidomic analysis reveals sphingomyelin and phosphatidylcholine species associated with renal impairment and all-cause mortality in type 1 diabetes. Sci. Rep. 9, 16398 (2019).
Lopes-Virella, M. F. et al. Glycosylated sphingolipids and progression to kidney dysfunction in type 1 diabetes. J. Clin. Lipido. 13, 481–491 e1 (2019).
Pongrac Barlovic, D. et al. Sphingomyelin and progression of renal and coronary heart disease in individuals with type 1 diabetes. Diabetologia 63, 1847–1856 (2020).
Kottgen, A. et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 42, 376–384 (2010).
Shiffman, D. et al. A gene variant in CERS2 is associated with rate of increase in albuminuria in patients with diabetes from ONTARGET and TRANSCEND. PLoS One 9, e106631 (2014).
Bellis, C. et al. Human plasma lipidome is pleiotropically associated with cardiovascular risk factors and death. Circ. Cardiovasc. Genet. 7, 854–863 (2014).
Aye, I. L., Rosario, F. J., Powell, T. L. & Jansson, T. Adiponectin supplementation in pregnant mice prevents the adverse effects of maternal obesity on placental function and fetal growth. Proc. Natl Acad. Sci. USA 112, 12858–12863 (2015).
Mahmoud, A. M. An overview of epigenetics in obesity: the role of lifestyle and therapeutic interventions. Int. J. Mol. Sci. 23, 1341 (2022).
Jin, Z. et al. Maternal adiponectin controls milk composition to prevent neonatal inflammation. Endocrinology 156, 1504–1513 (2015).
Okada-Iwabu, M. et al. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature 503, 493–499 (2013).
Freed, D., Aldana, R., Weber, J. A. & Edwards, J. S. The Sentieon Genomics Tools—a fast and accurate solution to variant calling from next-generation sequence data. bioRxiv, 115717 (2017) https://www.sentieon.com/citations/.
Hu, H. et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat. Biotechnol. 32, 663–669 (2014).
Pedersen, B. S. & Quinlan, A. R. Who’s who? Detecting and resolving sample anomalies in human dna sequencing studies with peddy. Am. J. Hum. Genet. 100, 406–413 (2017).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164–e164 (2010).
Singleton, M. V. et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Genet. 94, 599–610 (2014).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013).
Schraw, T., Wang, Z. V., Halberg, N., Hawkins, M. & Scherer, P. E. Plasma adiponectin complexes have distinct biochemical characteristics. Endocrinology 149, 2270–2282 (2008).
Lam, A. J. et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005–1012 (2012).
Acknowledgements
The authors would like to acknowledge grant support from the National Institutes of Health (DK47475 and DK58549 to A.S.K., DK108833 and DK112826 to W.L.H., DK115824, DK116888, and DK116450 to S.A.S., and DK128641 to M.G.P.), the National Institutes of Health’s and National Institute of Diabetes and Digestive and Kidney Disease’s Diabetes Research Center Program (P30 DK036836 to Joslin Diabetes Center), Diabetic Complications Consortium (DiaComp) (32307-1/U24-DK-115255 to M.G.P.) and Ruth L. Kirschstein National Research Service Awards (5T32DK091317 to C.A.S. and J.L.W.), the Juvenile Diabetes Research Foundation (JDRF 3-SRA-2019-768-A-B to S.A.S. and W.L.H.), the American Diabetes Association (to S.A.S.), the National Kidney Foundation of Utah & Idaho (M.G.P.), the Driving Out Diabetes, a Larry H. Miller Family Wellness Initiative (M.G.P.) and the Margolis Foundation (to S.A.S. and M.G.P.). We also acknowledge the Cell Imaging Core at the University of Utah for the use of equipment. Mass Spectrometry was performed at the Mass Spectrometry and Proteomics Core Facility at the University of Utah. Mass spectrometry equipment was obtained through a Shared Instrumentation Grant 1-S10-OD018210-01A1.
Author information
Authors and Affiliations
Contributions
A.S.K., W.L.H., and M.G.P. designed the study; C.A.S., J.L.W., W.L.H., and M.G.P. wrote the manuscript; C.A.S., J.L.W., A.M.P., J.A.B., J.V.V., J.L.G., E.J.H., B.G., D.L.A., T.T., S.G.F., J.C.F.M., B.M., M.Y., S.A.S., W.L.H., and M.G.P. analyzed the data and generated all the Figures and Tables; A.S.K. and K.O. provided DNA samples and clinical phenotype data; A.M.P., M.Y., and S.A.S. assisted in the data interpretation. All authors edited and have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.A.S. is a consultant, cofounder, and shareholder of Centaurus Therapeutics. The other authors declare no competing financial or nonfinancial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Simeone, C.A., Wilkerson, J.L., Poss, A.M. et al. A dominant negative ADIPOQ mutation in a diabetic family with renal disease, hypoadiponectinemia, and hyperceramidemia. npj Genom. Med. 7, 43 (2022). https://doi.org/10.1038/s41525-022-00314-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-022-00314-z