Abstract
New tools enable new ways of working, and materials science is no exception. In materials discovery, traditional manual, serial, and human-intensive work is being augmented by automated, parallel, and iterative processes driven by Artificial Intelligence (AI), simulation and experimental automation. In this perspective, we describe how these new capabilities enable the acceleration and enrichment of each stage of the discovery cycle. We show, using the example of the development of a novel chemically amplified photoresist, how these technologies’ impacts are amplified when they are used in concert with each other as powerful, heterogeneous workflows.
Similar content being viewed by others
Introduction
Events such as the COVID-19 global pandemic have starkly illustrated the need for ever accelerating cycles of scientific discovery. This challenge has instigated one of the greatest races in the history of scientific discovery—one that has demanded unprecedented agility and speed. This requirement is not localized to the healthcare domain, however; with significant pressure being exerted on the speed of materials discovery by challenges such as the climate emergency, which arguably are of an even greater magnitude. Fortunately, our tools for performing such discovery cycles are transforming—with data, artificial intelligence and hybrid cloud being used in new ways to break through long-standing bottlenecks1,2.
Historically, science has seen a number of major paradigm shifts, as depicted in Fig. 1, which have been driven by the advent and advancement of core underlying technology3. Moving from empirical studies, the collection and sharing of studies allowed a more global view of scientific problems, and led to the development of key underpinning theory. As the advent of computational systems allowed ever more complex calculations to be achieved, our understanding grew, with technology driving scale to new heights. The last two decades have seen the emergence of the Fourth Paradigm of big-data-driven science, dominated by an exa-flood of data4 and the associated systems and analytics to process it. The Fourth Paradigm has definitively made science a big-data problem5. For example, today virtual chemical databases contain billions of identified and characterized compounds6. Now, with the maturation of AI and robotic technology, alongside the further scaling of high-performance computing and hybrid cloud technologies, we are entering a new paradigm where the key is not any one individual technology, but instead how heterogeneous capabilities work together to achieve results greater than the sum of their parts.
A typical materials discovery effort can be decomposed into a series of phases: (1) specification of a research question, (2) collection of relevant existing data, then (3) formation of a hypothesis and finally (4) experimentation and testing of this hypothesis, which may in turn lead to knowledge generation and the creation of a new hypothesis. This process, whilst conceptually simple, has many significant bottlenecks which can hinder its successful execution7. For example, there are challenges in determining impactful research challenges, which requires increasingly deep and broad expertise. This is in part due to the known difficulties in keeping up with the rapidly expanding base of domain knowledge; for example, more than 28,000 articles were published on the subject of ‘photovoltaics’ since 2020—and this is just one area of research in our drive for renewable energy. Even when the materials space is constrained to be molecular, there are significant challenges in developing hypotheses relating structure to function, in part due to the sheer size of chemical space. Estimates say there are 10108 potential organic molecules8 which implies an intelligent navigation is necessary for any kind of accelerated discovery beyond serendipity. Similarly, there are gaps in experimentation, bridging digital models and physical testing, and ensuring reproducibility—it has been reported that 70% of scientists have at least once tried and failed to replicate the results of another9. Figure. 2 shows how the inclusion of AI, automation and improvements to deployment technologies can move towards a community-driven, closed loop process. This includes advances at each step, for example, to extract, integrate, and reason with knowledge at scale to better respond to question10, to the use of deep generative models to automatically propose new hypotheses, to automating testing and experimentation using robotic labs11. Important advances in the machine representation of knowledge also enable new results to lead to new questions and hypotheses12.
In this perspective, we describe technologies we have been exploring for this aim, and concretize our view with a real example where we have applied these technologies to a problem of commercial importance, the development of more sustainable photoacid generators (PAGs) for chemically amplified photoresists13.
Enabling technologies
Capturing unstructured technical data
Historical material science data is embedded in unstructured patents, papers, reports, and datasheets. Automated platforms are needed to ingest these documents, extract the data and ultimately present it to users for query and downstream use. For each individual component of the process, there are both proprietary and non-proprietary solutions available. For example, Semantic Scholar14 provides search access to over 190M scholarly articles. They also provide trained AI models for document conversion. Furthermore, solutions such as ChemDataExtractor15 and tmChem16 allow users to extract materials related entities from these documents, either in the generic case or for very specific use-cases—for example, zeolites17. Each of these components solve one aspect of the bigger problem, i.e. extracting unstructured materials related data from documents. We have devised the IBM DeepSearch platform to provide a holistic solution to the overall challenge of extracting unstructured data from documents, by having different tight-coupled services. These services allow users to upload documents and apply NLP algorithms on them in order to create KGs for deep queries (see Fig. 3).
Specifically, the IBM DeepSearch platform consists of the Corpus Conversion Service (CCS)18 and Corpus Processing Services (CPS)19. The CCS leverages state-of-the-art AI models20 to convert documents from PDF to the structured file format JSON. In this ingestion stage, there are a number of technical challenges—the segmentation of pages of the document into their component structure, the assignment of labels to each of these segments, and the identification and extraction of data from tables embedded in the document. To achieve sufficient accuracy across these tasks, a range of different models are required20,21,22. All these models run concurrently on a cloud-deployed cluster, enabling a conversion rate of 0.25 page/sec/core. This enables the conversion of the entire ArXiv repository23 in less than 24 h on 640 cores. Using the converted documents, the CPS service builds document-centric Knowledge Graphs (KGs) and supports rich queries and data extraction for downstream use. Common queries consist of searching for previously patented materials or associating reported properties with known materials. To this end, we pre-trained natural language processing (NLP) models for Named Entity Recognition (NER) of materials, properties, materials classes and unit-and-values. These entities become nodes in the graph linked by edges corresponding to detected relationships. Currently, the creation of KGs from corpora of hundreds of thousands of documents can be completed in approximately 6 h on 640 cores.
Key open challenges in the use of unstructured data in materials discovery include data access, entity resolution, and complex ad hoc queries. Data access is an issue as much of the content of interest, particularly technical papers and domain-specific databases, are not yet open access, particularly for large-scale machine ingestion. Navigating the appropriate copyright and usage agreements is often the most complex phase of an unstructured data project. The entity resolution problem in materials is often also complex. For example, the text of a paper might describe a material sample, which is then sub-divided and processed according to parameters shown in a table. Subsequent graphs of the properties for each individual sample may be labeled with symbolic references that require combining the information from both the text and the table to accurately identify the material and processing conditions which yield the graphed property. In effect, the materials entity is specified in a diffuse fashion across multiple modalities in the document. Finally, as capabilities to collect and organize materials data improve, there is a natural expectation that more complex queries should be supported, progressing from existence (‘Has this material been made?’) through performance (‘What’s the highest recorded Tc?’) to hypothesis (‘Could a Heusler compound be useful in this spintronic device?’).
Using AI to make simulation workflows more efficient and effective
The materials literature is overwhelmingly vast, but also incomplete24. Property data on existing materials is sparse, and data on hypothetical materials are necessarily absent. Simulation gives us the means to generate this data, but this switch from physical to digital experimentation provides some challenges. For example, the choice of simulation protocol can present complexity, and a poor choice can doom a discovery campaign before it is begun25. Even if an accurate protocol exists, the computational expense of executing it may severely limit the size of the design space being searched26. The area of AI or ML-assisted simulations can address some of these issues, and has been gaining some significant traction in recent years1,27. Emerging from the use of neural networks to bypass expensive physics-based routines28,29, AI has been used to predict ever more complex properties, such as energetic materials30, solid-state materials properties31,32 and even the structure of proteins33. In addition, we have seen the emergence of machine-learned potentials which enable access to quantum-chemical-like accuracies at a fraction of the cost34.
In our accelerated discovery paradigm, we consider that modeling and simulation workflows have the following structure.
-
Intent—the ‘translation’ of the property of interest into a corresponding computational workflow
-
Decision—the specific methodological choices used
-
Execution—scheduling, prioritization, and monitoring
-
Analysis—mapping output of the workflow to the real-world property
We believe that AI has the ability to enrich this structure in a number of ways. First, we perceive a valuable application of mature AI methodologies such as recommender systems to suggest which particular methodologies to use for particular tasks based on cost and accuracy. Where there is existing experimental data, this is trivial, but for new tasks, where data is sparse, this can become problematic. To circumvent this, we use a pairwise task similarity approach to guide the recommendation of low-data tasks from what we already know about high-data tasks. This method exploits the fact that the joint training of similar tasks will be broadly additive and positive, whilst the effect of joint training broadly dissimilar tasks will be net negative35. This has been shown to provide chemically plausible similarity measures for a range of tasks36.
Secondly, we can dynamically improve candidate prioritization using Bayesian optimization37,38, allowing us to selectively spend our computational budget, and thus use more accurate models on a smaller amount of data, thus improving the traditional ‘virtual high throughput screening’39 model shown in Fig. 4. This methodology is similar to other active learning approaches40 and allows us to balance the exploitation of trends from data we already have with the acquisition of new knowledge in unexplored areas41,42. Bayesian optimization is a general methodology often utilized when each data point is expensive (in time, cost, or effort) to acquire43. At each stage of screening, candidates are selected by optimizing an acquisition function which estimates the value for acquiring each data point. Improved Bayesian optimization algorithms allow the selection of batches of data points. Parallel Distributed Thompson Sampling37 parallelizes through sampling of the Bayesian model, while K-means Batch Bayesian optimization38 parallelizes through unsupervised partitioning of the acquisition function, and both have been deployed successfully to chemical discovery problems. In order to maximize the usability of this system, we present it to users through a simple set of cloud APIs known as IBM Bayesian Optimization (IBO).
a The ‘traditional’ computational funnel of high-throughput virtual screening. b Each level in the funnel is affected by the accuracy of screening, and the computational cost to perform the screen. Machine learning has the ability to markedly improve both of these, if the right training data is available.
The final part of the workflow where we believe AI can add value is to improve the relatedness of simulation outputs to real-world data. We achieve this through the calibration of the output of a simulation to better reflect an experimental outcome. This calibration is a type of delta machine learning44 with the addition of a concept of Bayesian uncertainty. Uncertainty aware models of how simulations systematically differ from experiment can provide highly accurate calibrations on a per-candidate basis, which avoid the pitfall of overfitting through the communication of a notion of the certainty of the correction (i.e. how much it can be trusted)25. The methodology we have chosen to achieve this goal is a Gaussian process model45 built on molecules described by their circular fingerprint46. This enables more robustness in the selection of simulation methods, and corresponding trust when making critical design decisions.
For these complex, AI-driven workflows to deliver on the promise of faster, more efficient simulation, several challenges need to be addressed. First, the practice of ‘virtual experiments’ that capture all aspects of a given computational task needs to become generally accepted and used in the community. Second, the traditional high performance computing model of manual or semi-manual long-running batch calculations will have to adapt to the dynamic active learning model described here. Finally, there is a need to eventually integrate the complex heterogeneous computing systems of the future, whether they are quantum computers, AI hardware accelerators, or classes of computer we cannot yet imagine.
Applying computational creativity to the molecular design problem
For molecular materials, across the wide range of structural scales, the molecular structure is dominant in determining properties of interest, and therefore, as we have previously noted, the materials design space can be intractably vast. In conventional molecular design processes, human experts explore this vast parameter space guided with their knowledge, experience, and intuition in a trial-and-error approach, which can yield a long development period and potentially limited variety. To counter this, we adopt an AI-driven generative modeling approach to collaborate with human experts and augment their creativity. Deep generative modeling (DGM) is one important example of such a class of technologies. Recent developments in AI technology based on pre-trained language models47 and Generative Adversarial Networks (GANs)48,49, have been used to automatically generate images, speech, and natural language, and have recently been applied to materials discovery problems50,51. In addition to DGM, other AI approaches have been effectively used for this purpose, including Monte Carlo tree search52, genetic algorithms53, and the junction tree algorithm54. Generative AI models can generate new candidate chemicals, molecules24,55,56, and materials57, and expand both the discovery space and the creativity of scientists. Our experience is that generative models can accelerate early materials ideation processes by 100x58.
Since there are a large number of approaches to generating materials candidates, it is important that the overall workflows are kept consistent. This can be distilled into the following common stages (see Fig. 5): after an initial training step, input molecular structures are encoded in a space that is used to predict associated properties. Next, the feature space (or latent space) is explored to sample feature vectors satisfying target properties. Finally, the sampled feature vectors are decoded to molecular structures. In deep generative models, our approach is to leverage Wasserstein Auto Encoders (WAE) and Conditional Latent Space Sampling (CLaSS) for this purpose which we have demonstrated successfully in peptide sequence generation to design antimicrobial materials59,60. Another approach is a combination of VAE and reinforcement learning (RL)61,62, where drug molecules’ SMILES and target proteins are both encoded on a common latent space. Reinforcement learning explores this space, guided by a model to predict the efficacy of the generated drug to target cancer proteins61. Another powerful scheme is the Molecular Generation Experience (MolGX), which leverages an explicit graph enumeration algorithm58,63. In MolGX, the encoder/decoder is pre-configured by the graph algorithms to generate valid chemical structures, therefore pre-training by a huge dataset is unnecessary. In addition, a user can fine tune the molecular generation process in atomistic detail (e.g. to control the number of (un)desired functional groups). The base MolGX functions are provided as a public web application at https://molgx.draco.res.ibm.com. The set of those generative models work under review and control by human experts, who tune and reinforce the models with domain knowledges.
Looking forward, generative models will need to evolve in their coverage of materials classes, extend beyond materials composition to processing and form, and effectively capture and encode application constraints based on human knowledge. The first of these, coverage of materials classes, is obvious, and will happen gradually as data becomes available. An open question is whether there will be a single unified generative model for all materials classes, or the gradual coverage of materials categories with independent models specialized for organic materials, crystalline inorganic materials, polymers, metal-organic frameworks, and so on. Regardless of the material category, these models will eventually need to capture the full complexity of materials manufacture and use. This will involve training the models on not just materials structures but also the un- and semi-structured data that describe materials synthesis and processing. Finally, we have found that in practice generative models are most useful when their outputs are either informed or filtered by the deep expertise of human subject matter experts. Tools and technologies to capture that expertise efficiently and encode it in the model will maximize the chances of generating not just a possible material, but a useful material.
Owing to its data-driven nature, generative model is highly compatible with open-innovative contribution by multiple users. For the sake of scalable offering of state-of-the-art pre-trained generative models and algorithms, we’ve been underway to integrate our algorithms onto a hybrid cloud platform, whereby users can contribute to the development and reuse them. We believe open science is a key concept to accelerate the evolution of generative modeling.
Materials evaluation using AI and cloud-powered automated labs
At the end of the design cycle, we face the need to accelerate the synthesis and testing of the large number of materials hypotheses. Recent advances in AI enabled digitization of common tasks in chemical synthesis, including forward reaction prediction64, retrosynthetic analysis65, and inference of experimental protocols to execute novel chemical synthesis66. Concurrently, there is an explosion of automation and AI in chemical synthesis, with the important contribution in the use of commodity hardware67, fluidic reactors68, or the use of robots able to execute the same tasks as human chemists69.
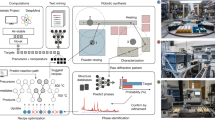
The construction of autonomous synthesis platforms is still a work in progress. One of the most recent efforts is RoboRXN, relying on an integration of three technologies: cloud, AI, and commercial automation to assist chemists all the way from the selection of synthetic routes to the actual synthesis of the molecule. A graphical overview of the architecture of RoboRXN is shown in Fig. 6. The use of cloud technologies enables a remote chemical laboratory as an embodiment of the cloud infrastructure, thus providing chemical services wherever an internet connection is available. AI is the core technology fueling the entire execution of domain experts’ tasks.
The prototype is made up of two parts. The first one which includes AI, Frontend and Backend components can live either in the Cloud or Premise thanks to the OpenShift technology that allows a seamless portability across different infrastructures. The second one comprising automation hardware physically located on the edge behind a firewall.
A core component of RoboRXN is a pipeline of multiple machine learning models that enables a complete automation of the synthesis plan, starting from a target molecule or a paragraph of a chemical recipe and ending with the process steps executed by the robot. Reaction prediction tasks are cast as translation tasks70, and trained on >2.5 million chemical reactions. These models, based on the Molecular Transformer64, allow the design of the synthesis starting from commercially available materials and provide an essential requirement of autonomous synthesis: the inference of precise sequence of operations that are executed by the synthesis hardware. All the models are freely available through a cloud platform called IBM RXN for Chemistry [https://rxn.res.ibm.com]. In the IBM lab, the integration of analytical chemistry technologies (LCMS and NMR) with the commercial synthesis hardware provides real-time monitoring to monitor results and generate feedback for improvement.
Future challenges in this space of automated chemistry include the generation and integration of in silico chemical data, the further integration of analytical chemistry and application-specific testing, and the expansion and adaptation of these technologies to other materials classes. The space of known chemical reactions, while vast, is still finite. In contrast, computational chemistry techniques could in principle allow the automated exploration of the vast space of hypothetical reactions. Intelligent exploration of this space could yield a wealth of training data for reaction prediction, provided it can be generated accurately and integrated with existing data-derived knowledge. These robotic systems also offer an opportunity for exploration of a different kind, specifically the prospect of independent active learning systems that on their own explore the chemical space searching for materials that suit application objectives. To do this effectively, the simulation and analytical chemistry systems need to be integrated with automated in-line application-specific testing. Finally, to fully realize the dream of a general-purpose materials synthesis robot, these capabilities need to be extended across materials classes. As with generative models, the question of whether there would ever be a single generalized robotic system or instead specialized robotic systems for specific materials and applications remains open.
Exemplar use case
As an exemplar use case, we carried out a project to address a key sustainability challenge focused on photoacid generators (PAGs), a critical photosensitive complex employed in chemically amplified photoresists used in semiconductor manufacturing71. Of the several classes of known PAGs72,73,74, sulfonium ([SR3]+) or iodonium ([IR2]+) -based complexes are the most widely used in semiconductor lithography75,76,77. Recently, onium-based photoacid generators have come under heightened regulatory scrutiny for potential persistence, bioaccumulation, and toxicity (PBT) risks78. While studies have helped clarify the potential PBT risks associated with representative PAGs79 as well as identify relevant photodecomposition products80,81, it remains extremely challenging for industry to design, synthesize, test, and bring to market new PAGs with improved sustainability profiles in a timely manner.
While both the cation and anion of prototypical onium PAGs could benefit from improved sustainability profiles, we initially focused our work on developing an accelerated discovery workflow for the discovery of sulfonium-based PAG cations with improved environmental health and safety profiles. The workflow is summarized in Fig. 7. In the first stage of the workflow, approximately 6000 patents, papers, and data sources were ingested by Deep Search to form a knowledge graph from which the structures of approximately 5000 sulfonium PAGs were obtained. To overcome the limited availability of key property values for most of the identified cations, AI-enriched simulation was used to compute both UV absorption using TD-DFT82 using the GAMESS-US framework83 and selected sustainability properties for several hundred sulfoniums using OPERA84. The predicted sustainability parameter set included basic physicochemical properties (octanol-water partition coefficient (LogP) and water solubility (LogWS)), an environmental persistence parameter (biodegradability—LogHalfLife), and a toxicity endpoint (CATMoS—LD50). The resulting structure-property dataset was then used to train a generative model, which was able to produce 3000 candidate sulfonium cations over the course of a 6 h run. However, many application-specific constraints that determine PAG utility in the context of semiconductor lithography were either partially or completely unaccounted for in the limited property dataset used to train the generative model. To overcome this, a combination of expert-defined rules and discriminative expert-in-the-loop (EITL) AI models85 were used to first constrain the generative model output and then aid in the candidate downselection process, respectively. In this manner, the 3000 PAG candidates output from the generative model were filtered down to only a few hundred candidates, a more manageable number for which property data could be simulated.
Simulation on these candidates was prioritized using the aforementioned IBM Bayesian optimization (IBO) functionality, using a simple scoring metric, S, which combined the distance from the target excitation energy (6.46 eV and 5.00 eV for the common 193 nm and 248 nm targets) and the computed oscillator strength:
where Eobs is the computed excitation energy, Etarget is the target excitation energy and fobs is the computed oscillator strength.
An example of the speedup possible through this method can be seen in Fig. 8, which shows a comparison between an IBO accelerated workflow, and a non-accelerated workflow. For this example, IBO was configured to use PDTS (parallel, distributed Thompson sampling37), collecting batches of 10 simulations in parallel, with molecules described using ECFP descriptors46. ECFP fingerprints were chosen due to their previous successes at this task37,41, their speed of calculation relative to other ‘learned’ representations, and their ability to be generated from 2D information. For a library of over 400 candidate PAGs, the highest performing molecule targeting the 193 nm wavelength was found on average after only testing half the library, with the best performing accelerated workflow locating this candidate after only testing 90 molecules.
Solid lines represent bootstrap estimates of the mean run from 5 replicate workflows, with shaded areas representing 95% confidence intervals for that estimate. Ideal behavior (i.e. the best possible score) is shown as a gray dashed line, and the best IBO-accelerated workflow is shown as a blue dashed line.
This multi-step refinement methodology enabled a 100-fold reduction in the number of generated candidates that a human environmental toxicology expert was required to analyze in order to perform a final selection of a few top candidates for the next stage of automated retrosynthetic analysis. With several top candidates consisting of substituted variants of a dialkylphenylsulfonium core, (4-methylphenyl)dimethylsulfonium triflate was employed as a model candidate surrogate to simplify the final experimental validation. Application of the AI retrosynthetic model identified a one-step reaction involving the S-alkylation of 4-(methylthio)toluene by methyl trifluoromethanesulfonate86,87 as the most promising pathway (shown in Fig. 9). The reaction instruction set was generated and transferred via the cloud to the RoboRXN system, which successfully carried out the reaction to afford the expected product. This initial demonstration of the applicability and utility of the discovery workflows for PAGs has inspired us to begin expanding the simulation portfolio and diversifying the types of generative AI models used in the workflow for future discovery cycles.
Outlook
The work described above illustrates a prototype for the future of accelerated materials discovery. There are certainly examples of larger-scale computational screening efforts88, as well as more complex laboratory automation69, but in contrast, the workflow we describe here is much more irregular and heterogeneous, requiring the linking together of multiple distinct capabilities over multiple geographies. Of note, the complexity of this irregular and heterogeneous discovery workflow was enabled by the use of the OpenShift hybrid cloud computing framework89, enabling a single researcher to orchestrate the available resources across 3 data-centers on 3 continents to execute the necessary steps – a model which we believe will become more essential as the task of materials discovery continues to globalize and new technologies such as quantum computing continue to challenge what is possible in each stage of the discovery cycle. In this prototype, a series of sophisticated applications, algorithms, and computational systems are seamlessly orchestrated to accelerate cycles of learning and support human scientists in their quest for knowledge. In our research, we have seen tangible examples of this acceleration across all stages of the discovery process, and we strongly believe that the commoditization and democratization of such diverse workflows will fundamentally alter the way we respond to emerging discovery challenges.
References
Suh, C., Fare, C., Warren, J. A. & Pyzer-Knapp, E. O. Evolving the materials genome: how machine learning is fueling the next generation of materials discovery. Annu. Rev. Mater. Res. 50, 1–25 (2020).
Tabor, D. P. et al. Accelerating the discovery of materials for clean energy in the era of smart automation. Nat. Rev. Mater. 3, 5–20 (2018).
Kuhn, T. The Structure of Scientific Revolutions 2nd edn (The University of Chicago Press, 1970).
Leonelli, S. Scientific Research and Big Data (Stanford Encyclopedia of Philosophy, 2020).
Hey, A. J., Tansley, S., Tolle, K. M. The Fourth Paradigm: Data-intensive Scientific Discovery 1 (Microsoft Research Redmond, 2009).
Ruddigkeit, L., van Deursen, R., Blum, L. C. & Reymond, J.-L. Enumeration of 166 Billion Organic Small Molecules in the Chemical Universe Database GDB-17. J. Chem. Inf. Model. 52, 2864–2875 (2012).
Zubarev, D. Y. & Pitera, J. W. In Machine Learning in Chemistry: Data-Driven Algorithms, Learning Systems, and Predictions (eds Pyzer-Knapp, E. O. & Laino, T.) 103–120 (ACS Publications, 2019).
Reymond, J.-L., Ruddigkeit, L., Blum, L. & van Deursen, R. The enumeration of chemical space. WIREs Comput. Mol. Sci. 2, 717–733 (2012).
Baker, M. Reproducibility crisis. Nature 533, 353–66 (2016).
Spangler, S. et al. Automated hypothesis generation based on mining scientific literature. In: Proc. 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 1877–1886 (2014).
Vaucher, A. C. et al. Automated extraction of chemical synthesis actions from experimental procedures. Nat. Commun. 11, 1–11 (2020).
Kohli, P. AI Will Help Scientists Ask More Powerful Questions. Scientific American Blog Network https://blogs.scientificamerican.com/observations/ai-will-help-scientists-ask-more-powerful-questions/.
Willson, C. G., Ito, H., Fréchet, J. M., Tessier, T. G. & Houlihan, F. M. Approaches to the design of radiation-sensitive polymeric imaging systems with improved sensitivity and resolution. J. Electrochem. Soc. 133, 181 (1986).
Lo, K., Wang, L. L., Neumann, M., Kinney, R. & Weld, D. S2ORC: The Semantic Scholar Open Research Corpus. In: Proc. 58th Annual Meeting of the Association for Computational Linguistics 4969–4983 (Association for Computational Linguistics, 2020).
Swain, M. C. & Cole, J. M. ChemDataExtractor: a toolkit for automated extraction of chemical information from the scientific literature. J. Chem. Inf. Model. 56, 1894–1904 (2016).
Leaman, R., Wei, C.-H. & Lu, Z. tmChem: a high performance approach for chemical named entity recognition and normalization. J. Cheminformatics 7, S3 (2015).
Jensen, Z. et al. A machine learning approach to zeolite synthesis enabled by automatic literature data extraction. ACS Cent. Sci. 5, 892–899 (2019).
Staar, P. W. J., Dolfi, M., Auer, C. & Bekas, C. Corpus conversion service: a machine learning platform to ingest documents at scale. In: Proc. 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining 774–782 (ACM, 2018).
Staar, P. W. J., Dolfi, M. & Auer, C. Corpus processing service: a knowledge graph platform to perform deep data exploration on corpora. Appl AI Lett. 1, e20 (2020).
Livathinos, N. et al. Robust PDF document conversion using recurrent neural networks. Proc. AAAI Conf. Artif. Intell. 35, 15137–15145 (2021).
Wu, Y., Kirillov, A., Massa, F., Lo, W.-Y. & Girshick, R. https://github.com/facebookresearch/detectron2. (2019).
Zhong, X., ShafieiBavani, E. & Jimeno Yepes, A. In Computer Vision—ECCV 2020 (eds. Vedaldi, A., Bischof, H., Brox, T. & Frahm, J.-M.) 564–580 (Springer International Publishing, 2020).
arXiv.org e-Print archive. https://arxiv.org/.
Himanen, L., Geurts, A., Foster, A. S. & Rinke, P. Data-driven materials science: status, challenges, and perspectives. Adv. Sci. 6, 1900808 (2019).
Pyzer-Knapp, E. O., Simm, G. N. & Guzik, A. A. A Bayesian approach to calibrating high-throughput virtual screening results and application to organic photovoltaic materials. Mater. Horiz. 3, 226–233 (2016).
Pyzer-Knapp, E. O., Chen, L., Day, G. M. & Cooper, A. I. Accelerating computational discovery of porous solids through improved navigation of energy-structure-function maps. Sci. Adv. 7(33), eabi4763 (2021).
Cartwright, H. M. Machine Learning in Chemistry. (Royal Society of Chemistry, 2020).
Montavon, G. et al. Machine learning of molecular electronic properties in chemical compound space. N. J. Phys. 15, 095003 (2013).
Pyzer‐Knapp, E. O., Li, K. & Aspuru‐Guzik, A. Learning from the Harvard clean energy project: the use of neural networks to accelerate materials discovery. Adv. Func. Mater. 25.41, 6495–6502 (2015).
Elton, D. C., Boukouvalas, Z., Butrico, M. S., Fuge, M. D. & Chung, P. W. Applying machine learning techniques to predict the properties of energetic materials. Sci. Rep. 8, 9059 (2018).
Goodall, R. E. A. & Lee, A. A. Predicting materials properties without crystal structure: deep representation learning from stoichiometry. Nat. Commun. 11, 6280 (2020).
Schmidt, J., Marques, M. R. G., Botti, S. & Marques, M. A. L. Recent advances and applications of machine learning in solid-state materials science. npj Comput Mater. 5, 1–36 (2019).
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017).
Xu, Y., Ma, J., Liaw, A., Sheridan, R. P. & Svetnik, V. Demystifying multitask deep neural networks for quantitative structure–activity relationships. J. Chem. Inf. Model. 57, 2490–2504 (2017).
Fare, C., Turcani, L. & Pyzer-Knapp, E. O. Powerful, transferable representations for molecules through intelligent task selection in deep multitask networks. Phys. Chem. Chem. Phys. 22.23, 13041–13048 (2020).
Hernández-Lobato, J. M., Requeima, J., Pyzer-Knapp, E. O. & Aspuru-Guzik, A. Parallel and distributed Thompson sampling for large-scale accelerated exploration of chemical space. In: Proc. 34th International Conference on Machine Learning-Volume 70 1470–1479 (JMLR. org, 2017).
Groves, M. & Pyzer-Knapp, E. O. Efficient and Scalable Batch Bayesian Optimization Using K-Means. Preprint at https://arxiv.org/abs/1806.01159 (2018).
Pyzer-Knapp, E. O., Suh, C., Gómez-Bombarelli, R., Aguilera-Iparraguirre, J. & Aspuru-Guzik, A. What Is high-throughput virtual screening? a perspective from organic materials discovery. Annu. Rev. Mater. Res. 45, 195–216 (2015).
Kusne, A. G. et al. On-the-fly closed-loop materials discovery via Bayesian active learning. Nat. Commun. 11, 5966 (2020).
Pyzer-Knapp, E. Bayesian optimization for accelerated drug discovery. IBM J. Res Dev. 62, 2–1 (2018).
Pyzer-Knapp, E. O. Using Bayesian optimization to accelerate virtual screening for the discovery of therapeutics appropriate for repurposing for COVID-19. Preprint at https://arxiv.org/abs/2005.07121 (2020).
Brochu, E., Cora, V. M. & de Freitas, N. A Tutorial on Bayesian optimization of expensive cost functions, with application to active user modeling and hierarchical reinforcement learning. Preprint at https://arxiv.org/abs/10122599 (2010).
Ramakrishnan, R., Dral, P. O., Rupp, M. & von Lilienfeld, O. A. Big data meets quantum chemistry approximations: the Δ-machine learning approach. J. Chem. Theory Comput. 11, 2087–2096 (2015).
Rasmussen, C. E. Gaussian Processes for Machine Learning. (MIT Press, 2006).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Brown, T. B. et al. Language models are few-shot learners. Preprint at https://arxiv.org/abs/2005.14165 (2020).
Guimaraes, G. L., Sanchez-Lengeling, B., Outeiral, C., Farias, P. L. C. & Aspuru-Guzik, A. Objective-reinforced generative adversarial networks (ORGAN) for sequence generation models. Preprint at https://arxiv.org/abs/1705.10843 (2018).
Maziarka, Ł. et al. Mol-CycleGAN: a generative model for molecular optimization. J. Cheminformatics 12, 2 1–18 (2020).
Gómez-Bombarelli, R. et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018).
Kusner, M. J., Paige, B. & Hernández-Lobato, J. M. Grammar variational autoencoder. In: International Conference on Machine Learning 1945–1954 (PMLR, 2017).
Yang, X., Zhang, J., Yoshizoe, K., Terayama, K. & Tsuda, K. ChemTS: an efficient python library for de novo molecular generation. Sci. Technol. Adv. Mater. 18, 972–976 (2017).
Jensen, J. H. A graph-based genetic algorithm and generative model/Monte Carlo tree search for the exploration of chemical space. Chem. Sci. 10, 3567–3572 (2019).
Jin, W., Barzilay, R. & Jaakkola, T. Junction tree variational autoencoder for molecular graph generation. In: International Conference on Machine Learning 2323–2332 (PMLR, 2018).
Jørgensen, P. B., Schmidt, M. N. & Winther, O. Deep generative models for molecular science. Mol. Inf. 37, 1700133 (2018).
Schwalbe-Koda, D. & Gómez-Bombarelli, R. Generative models for automatic chemical design. In Machine Learning Meets Quantum Physics. pp. 445–467 (Springer, Cham, 2020).
Maziarka, Ł. et al. Molecule attention transformer. Preprint at https://arxiv.org/abs/arXiv:2002.08264 (2020).
Takeda, S. et al. Molecular inverse-design platform for material industries. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (2020).
Das, P. et al. PepCVAE: semi-supervised targeted design of antimicrobial peptide sequences. Preprint at https://arxiv.org/abs/1810.07743 (2018).
Das, P. et al. Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat. Biomed. Eng. 5, 613–623 (2021).
Manica, M. et al. Toward explainable anticancer compound sensitivity prediction via multimodal attention-based convolutional encoders. Mol. Pharmaceutics 16, 4797–4806 (2019).
Cadow, J., Born, J., Manica, M., Oskooei, A. & Rodríguez Martínez, M. PaccMann: a web service for interpretable anticancer compound sensitivity prediction. Nucleic Acids Res. 48, W502–W508 (2020).
Takeda, S. et al. Molecule generation experience: an open platform of material design for public users. Preprint at https://arxiv.org/abs/2108.03044 (2021).
Schwaller, P. et al. Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Cent. Sci. 5(9), 1572–1583 (2019).
Schwaller, P. et al. Predicting retrosynthetic pathways using transformer-based models and a hyper-graph exploration strategy. Chem. Sci. 11, 3316–3325 (2020).
Vaucher, A. C. et al. Inferring experimental procedures from text-based representations of chemical reactions. Nat. Commun. 12, 2573–11 (2021).
Angelone, D. et al. Convergence of multiple synthetic paradigms in a universally programmable chemical synthesis machine. Nat. Chem. 13, 63–69 (2021).
Coley, C. W. et al. A robotic platform for flow synthesis of organic compounds informed by AI planning. Science 365, 6453 (2019): eaax1566 (2019).
Burger, B. et al. A mobile robotic chemist. Nature 583, 237–241 (2020).
Schwaller, P., Gaudin, T., Lányi, D., Bekas, C. & Laino, T. ‘Found in Translation’: predicting outcomes of complex organic chemistry reactions using neural sequence-to-sequence models. Chem. Sci. 9, 6091–6098 (2018).
Ito, H. In Microlithography. Molecular Imprinting 37–245 (Springer Berlin Heidelberg, 2005).
Tsuchimura, T. Recent progress in photo-acid generators for advanced photopolymer materials. J. Photopolym. Sci. Technol. 33, 15–26 (2020).
Kuznetsova, N. A., Malkov, G. V. & Gribov, B. G. Photoacid generators. Application and current state of development. Russ. Chem. Rev. 89, 173–190 (2020).
Zivic, N. et al. Recent advances and challenges in the design of organic photoacid and photobase generators for polymerizations. Angew. Chem. Int. Ed. 58, 10410–10422 (2019).
Crivello, J. V. The discovery and development of onium salt cationic photoinitiators. J. Polym. Sci. A Polym. Chem. 37, 4241–4254 (1999).
Crivello, J. V. & Lam, J. H. W. Photoinitiated cationic polymerization with triarylsulfonium salts. J. Polym. Sci. A: Polym. Chem. 17, 977–999 (1979).
Crivello, J. V. & Lam, J. H. W. Diaryliodonium salts. a new class of photoinitiators for cationic polymerization. Macromolecules 10, 1307–1315 (1977).
Tvermoes, B. & Speed, D. Increased regulatory scrutiny of photolithography chemistries: the need for science and innovation (Conference Presentation). In: Advances in Patterning Materials and Processes XXXVI (eds. Gronheid, R. & Sanders, D. P.) (SPIE, 2019). https://doi.org/10.1117/12.2516159.
Niu, X.-Z. et al. Bioconcentration potential and microbial toxicity of onium cations in photoacid generators. Environ 28, 8915–8921 (2021).
Niu, X.-Z. et al. Photochemical fate of sulfonium photoacid generator cations under photolithography relevant UV irradiation. J. Photochem. Photobiol. A 416, 113324 (2021).
Despagnet-Ayoub, E. et al. Triphenylsulfonium topophotochemistry. Photochem. Photobiol. Sci. 17, 27–34 (2018).
Runge, E. & Gross, E. K. Density-functional theory for time-dependent systems. Phys. Rev. Lett. 52, 997 (1984).
Barca, G. M. J. et al. Recent developments in the general atomic and molecular electronic structure system. J. Chem. Phys. 152, 154102 (2020).
Mansouri, K., Grulke, C. M., Judson, R. S. & Williams, A. J. OPERA models for predicting physicochemical properties and environmental fate endpoints. J. Cheminformatics 10, 10 (2018).
Ristoski, P. et al. Expert-in-the-loop AI for polymer discovery. In: Proc. 29th ACM International Conference on Information & Knowledge Management (ACM, 2020). https://doi.org/10.1145/3340531.3416020.
Minami, H., Otsuka, S., Nogi, K. & Yorimitsu, H. Palladium-catalyzed borylation of aryl sulfoniums with diborons. ACS Catal. 8, 579–583 (2017).
Huang, C. et al. Redox-neutral borylation of aryl sulfonium salts via C–S activation enabled by light. Org. Lett. 21, 9688–9692 (2019).
Carrete, J., Li, W., Mingo, N., Wang, S. & Curtarolo, S. Finding unprecedentedly low-thermal-conductivity half-Heusler semiconductors via high-throughput materials modeling. Phys. Rev. X 4, 011019 (2014).
Shipley, G. & Dumpleton, G. OpenShift for Developers: A Guide for Impatient Beginners (O’Reilly Media, Inc., 2016).
Acknowledgements
The authors would like to acknowledge their IBM colleagues for their continued contributions to this exciting area of research. In particular, we would like to thank Dmitry Zubarev and Brooke Tvermoes for helpful conversations during the preparation of this perspective.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception, structuring, and writing of this perspective.
Corresponding author
Ethics declarations
Competing interests
The authors declare no Competing Non-Financial Interests but declare the following Competing Financial Interests: All authors are employed by IBM Research, which has an active research program in accelerated discovery. The IBM DeepSearch, IBM Bayesian Optimization, IBM Molecule Generation Experience and IBM RoboRXN technologies described in this Perspective were developed as part of that effort.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pyzer-Knapp, E.O., Pitera, J.W., Staar, P.W.J. et al. Accelerating materials discovery using artificial intelligence, high performance computing and robotics. npj Comput Mater 8, 84 (2022). https://doi.org/10.1038/s41524-022-00765-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-022-00765-z
This article is cited by
-
Embracing data science in catalysis research
Nature Catalysis (2024)
-
GEMTELLIGENCE: Accelerating gemstone classification with deep learning
Communications Engineering (2024)
-
Rapid and accurate predictions of perfect and defective material properties in atomistic simulation using the power of 3D CNN-based trained artificial neural networks
Scientific Reports (2024)
-
A dynamic knowledge graph approach to distributed self-driving laboratories
Nature Communications (2024)
-
An active machine learning approach for optimal design of magnesium alloys using Bayesian optimisation
Scientific Reports (2024)