Abstract
Empirical interatomic potentials require optimization of force field parameters to tune interatomic interactions to mimic ones obtained by quantum chemistry-based methods. The optimization of the parameters is complex and requires the development of new techniques. Here, we propose an INitial-DEsign Enhanced Deep learning-based OPTimization (INDEEDopt) framework to accelerate and improve the quality of the ReaxFF parameterization. The procedure starts with a Latin Hypercube Design (LHD) algorithm that is used to explore the parameter landscape extensively. The LHD passes the information about explored regions to a deep learning model, which finds the minimum discrepancy regions and eliminates unfeasible regions, and constructs a more comprehensive understanding of physically meaningful parameter space. We demonstrate the procedure here for the parameterization of a nickel–chromium binary force field and a tungsten–sulfide–carbon–oxygen–hydrogen quinary force field. We show that INDEEDopt produces improved accuracies in shorter development time compared to the conventional optimization method.
Similar content being viewed by others
Introduction
Atomistic-scale insights have been critical to understanding the dynamical evolution of chemically reactive systems and therefore have created a demand for the development of computational chemistry techniques. Quantum mechanics (QM)-based atomistic simulation methods have been commonly used to study the molecular dynamics (MD) because these methods provide accurate energies, charges, and reaction pathways. However, simulation times and sizes are highly constrained by computational cost, and these limitations are some of the motivations behind the development of empirical potentials. The empirical potentials provide fast access to forces and, in turn, the dynamical evolution of much larger chemical systems for longer simulation times in various conditions (e.g., temperature, pressure). However, due to fixed connectivity between atoms, traditional empirical potentials have limited capability of modeling the evolution of systems during reactive events. In order to bridge the gap between QM and empirical potential-based methods, several reactive force field-based atomistic simulation techniques have been developed1,2,3,4. ReaxFF is one of the widely used reactive interatomic potential in this category due to its reliability and transferability between chemical systems. Initially, ReaxFF interatomic potential has been formulized to model hydrocarbons, and then extended through silica, nitramine-based materials to several aqueous and combustion systems5. Currently, the ReaxFF method covers over 50 elements, and is applicable to a wide range of chemical systems that are of interest to materials science community (i.e., 2D materials6,7,8,9,10,11,12,13, electrochemistry14,15, thin films16,17, nanotubes18,19, catalysts20,21).
ReaxFF uses bond-order and charge-dependent functionals to define intra- and interatomic interactions. The simplest form of the total energy equation used in ReaxFF can be given as below, and a detailed and most up-to-date form can be found in ref. 22:
where Ebond, Eangle, and Etors are energy terms related to bond formation, three-body valence angle strain, and four-body torsional angle strain, respectively. The energy term, Eover, is used to prevent the over coordination of atoms. The energy terms, ECoulomb and EvdW, are associated with electrostatic and dispersive contributions, respectively. Each separate energy term in Eq. (1) contributes to the total energy of the system, which in turn predicts the dynamical evolution. Each term is calculated by an equation involving several parameters to be optimized to adapt the calculation to a form specific to a chemical system of interest. A typical ReaxFF force field consists of approximately 100 of these parameters per element type depending on the element. Because the intra- and interatomic interactions are tuned by these parameters, they are optimized to reproduce reference values with reasonable accuracy before moving to production simulations. These reference values form a force field training (FF-training) set, which is composed of, but not limited to, molecular properties (e.g. bond lengths, bond angles, charges, and energies), and/or chemical reactions from as simple as bond breaking/formation to more complex such as vacancy dynamics, ion diffusion of reference systems. The reference values, which hereafter will be referred to as “reference properties”, are obtained using QM-based methods if experimental values are not present.
The quality of the production simulations is strictly affected by the quality of the developed force field; therefore, the force field parametrization is one of the most critical parts of the MD studies. ReaxFF uses fixed functions for all chemical systems; hence, application to different chemical systems and/or different applications of similar chemical systems require adaptation of ReaxFF through optimization of the force field parameters. The optimization process requires a comprehensive exploration of the force field parameter landscape. However, optimization of such a large number of parameters, especially for multi-component chemical systems, is challenging to achieve. A typical force field for a multi-component system contains several hundreds of parameters, a significant part of which can be transferred from previously optimized force fields; however, depending on the complexity of the molecular system, a few tens of parameters should be optimized to reproduce reference properties in a given FF-training set. A parameter space that is composed of tens of parameters can be considered as high-dimensional and can have a complex geometry with many local minima.
Each local minimum in ReaxFF parameter space yields acceptable molecular property values and therefore can be reliably used by researchers for the application of the developed force field in the production stage. However, being stuck in a local minimum may be troublesome from the parametrization perspective. For example, it is a very common practice to update FF-training sets to extend the applications, and this update requires reparameterization of the force field because the previously found local minimum corresponding to an ideal parameter set may no longer be appropriate for newly added reference properties. Another issue that may arise during force field development is due to diversity in the FF-training set. A typical ReaxFF training set is composed of molecular and condensed phase parts, which are also separated into groups such as the equation of states, the heat of formations, etc. It is very likely for a force field parameter set to get stuck in a local minimum that precisely reproduce reference properties corresponding to a specific part of the force field while poorly fitting to the remaining.
The ability to have the information of several local minima in parameter space and so making transitions between these minima is significantly beneficial to detect low discrepancy regions that are capable of precisely fitting to the whole FF-training set, including all sections. The conventional optimization method that is widely used by the force field developers is a sequential one parameter parabolic extrapolation method23,24, which is not capable of switching between local minima to detect lowest error regions in parameter space. In contrast, this method is susceptible to being stuck in a local minimum due to its sequentiality, which also prevents parallelization of the optimization algorithm. Due to these limitations in the conventional method, considerable effort has been directed toward finding a solution to this optimization problem25,26,27,28,29,30. Iype et al.25 have used a Monte Carlo algorithm combined with simulated annealing (SA) procedure to find global optimum values for ReaxFF parameters of magnesium-salt hydrates. The application of SA method varies depending on the system; therefore, it cannot offer a common solution to the force field optimization problem. Guo et al.31 developed a method to improve ReaxFF parameterization using machine learning and fit separate ReaxFF energy terms (Eq. 1) to reproduce density functional theory (DFT) MD energy terms with high accuracy; however, the applications of this method were suitable for very small three-component chemical systems trained over a few single phases. Several studies have used single and multi-objective global optimization methods such as genetic algorithm (GA)26,27,28,29,30 or enhanced particle swarm optimization method32 to optimize ReaxFF force field parameters. All these studies have implemented existing global optimization methods to parameterize ReaxFF force fields and despite how useful these methods have become; the optimization problem has not yet been completely solved. However, we believe that the ReaxFF optimization system should not be viewed as “just another” global optimization problem, because there may be several parameter combinations that may be “almost equally good” or” equally desirable”.
Here we present an INitial-DEsign Enhanced Deep learning-based OPTimization (INDEEDopt) framework that will not only accelerate the force field development for simple chemical systems (e.g., ternary, quaternary, or quinary component), but also enable the development for multi-component ones (e.g., senary or larger), which have become crucial with the advances in materials discovery (e.g., high entropy alloys, inorganic–organic interfaces). The framework uses machine learning (ML) due to its capability of modeling highly correlated high-dimensional data and adapts an initial design algorithm to explore the high-dimensional parameter space comprehensively. Thus, INDEEDopt is capable of finding several local minima in parameter space and produces optimized force fields to be used in the production stage of simulations. INDEEDopt differs from other interesting ReaxFF parameterization methodologies in several aspects. An important feature of INDEEDopt is that it uses a Latin Hyper Design (LHD) algorithm to efficiently generate a data set and explore whole parameter space instead of being stuck around initially assigned regions. Most of the optimization methods developed to date require initial parameter value guesses; however, there is no certain rule to assign these values. INDEEDopt does not require the initial values; in fact, it can be used as a method to generate these values that can be further optimized by combining with other optimization methods. Another critical distinction is that INDEEDopt is a data-driven method and generates a significant amount of data, which can be used for further investigations. The procedure that INDEEDopt framework uses is fully parallelizable and can be used with any ReaxFF-MD software without modifications to the code. INDEEDopt as a concept can be adapted to the parametrization of classical and other reactive force fields in the future if desired.
Results
INDEEDopt force field optimization framework
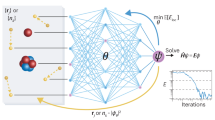
INDEEDopt framework involves three stages, and a schematic representation of the framework can be seen in Fig. 1. In the first stage, the high-dimensional parameter space is sampled by means of an initial design algorithm called Orthogonal-maximin Latin Hypercube Design (OMLHD). The OMLHD algorithm can generate parameter combinations within ranges specific to each parameter that are multidimensionally uniformly distributed by reducing the pairwise correlation and maximizing the distance between parameters33. The parameter combinations are given to the ReaxFF-MD code for energy minimization calculations, which returns target property values correspond to each reference property in FF-training set. The parameter combinations and corresponding target property values produce a training set for the second stage of the framework, which is the ML model training stage, and hereafter as this training set will be referred to as ML-training set.
Schematic representation of the DL-based optimization procedure. a Representation of the sampling using the initial design algorithm. b Deep neural network structure, red circles are input nodes, yellow circles are output nodes, and grey circles are hidden layer nodes. c Representation of the parameter-discrepancy correlation in parameter space.
The strong correlations of ReaxFF force field parameters create very complex parameter–property relationships because these correlations do not only come from analytical expressions but are also correlated due to the many-body character of atomic interactions specific to the system. In order to handle these complex correlative relationships, we adapted deep learning (DL) as the ML method in the framework, because DL is a promising approach in extracting high-level features from less featured input data34. The DL involves a type of artificial neural network with multiple hierarchical hidden layers. Using these multi-layered network structures, DL models are capable of nonlinearly mapping the relationships between a given set of inputs and corresponding outputs. This capability makes DL an appropriate method to obtain features from a large data set. In addition, DL models can be easily parallelized on CPU or GPU architectures, which is one of the goals of this study since this provides significant acceleration in the FF development.
In the DL model, the ReaxFF parameters (N) are given as an input to a deep neural network (DNN), which is trained to return target molecular property values (P) (Fig. 1b). The DNN used in this study is a fully connected feed-forward type, which transfers data from the input to the output layer through hidden layers. Each layer is composed of several nodes that are connected to each of the other nodes in the previous and next layers. Nonlinear mapping is implemented into neural networks using activation functions. For all layers except the last layer, rectified linear unit (ReLu), which has been proven to accelerate the training process35, is employed as an activation function, and we use a linear activation function for the last layer. The shape of a neural network used in DL model affects prediction accuracy36; therefore, to understand the complexity of the neural network used in INDEEDopt, we analyzed the effect of the network shape to the final prediction. The accuracy of prediction was measured according to the mean absolute error (MAE) between the model prediction and test data. According to our analysis, MAE decreases until the number of layers is increased over ten. After ten, the model tends to overfit to the data, thus results in a reduction in accuracy (Fig. 2). We used two hidden layers in our model because the accuracy difference between ten and two layers is not significant enough to negatively affect the optimization procedure. The effect of the number of nodes per hidden layer was analyzed by changing the node amount in each layer by multiples of the number of reference property values in the FF-training set. As can be seen from Fig. 2, the increase in the number of nodes results in a decrease in MAE, and the network with 900 nodes per layer fits best to the test data. However, the improvement in accuracy is not significant, while the increase in node number is inefficient in terms of computational time. In the light of these analyses, two hidden layers containing a constant number of nodes, which was selected as the size of the FF-training set, is considered as the most appropriate neural network configuration to be used in INDEEDopt framework. For a more complex FF-training set that is composed of thousands of reference property values, there may be a need to assign more hidden layers and more nodes per layer; however, our configuration produced sufficient accuracy for both of our parametrization test cases and should be sufficient for most of the parametrizations conducted in current ReaxFF literature. The INDEEDopt code is capable of detecting GPU architecture in the computer system and automatically run on it, so it is advisable to use a larger number of nodes and layers if GPU is accessible to force field developer.
The effect of neural network shape on the accuracy of the DL model. All model training was run until the mean absolute error (MAE) is converged. The change in MAE with respect to the number of hidden layers (a) and the effect of number of nodes per layer to MEA of the trained model (b). The error bars were calculated training each model on the same data with ten repetitions. The error bars represent standard deviation (s.d.). The training data output values (i.e., property values) are standardized to have a mean of zero and a standard deviation of one.
The DL training loss function is optimized using Adaptive Moment Estimation (Adam) method, which is computationally efficient and suitable for high dimensionality due to its adaptive learning rate37. Once the DL model is trained, the accuracy of the model is evaluated using a test set, which is obtained by randomly selecting a part of the ML-training set as a test set with a split ratio of 0.2. The DL model has an early stopping algorithm, which ensures that the overfitting is avoided by stopping the training at the point where the prediction accuracy is maximum. The maximum fail parameter is set to 100 epochs, while the total number of epochs is selected as 10,000. The quality of the DL model predictions is calculated using MAE. The first two stages of INDEEDopt are iterative and repeat until the DL model converges to an acceptable accuracy. The accuracy is calculated by the MAE between DL prediction and ReaxFF calculation. The third stage of the framework, which is the optimization stage, starts once the accuracy is converged. The convergence criterion is satisfied when the accuracy change between two consecutive iterations is less than 2%. The accuracy convergence plots for two different applications of INDEEDopt can be seen in Supplementary Fig. 1. The computational flow diagram of INDEEDopt framework is depicted in Fig. 3. We should note that the convergence of DL model is critical, and the third stage does not start until the convergence in second stage is achieved. A well converged DL model is capable of reproducing ReaxFF calculations with high accuracy as can be seen in Fig. 4, which shows how close are the predictions and the true values. The percent difference between the INDEEDopt predictions and ReaxFF calculations can also be seen in Supplementary Table 1. There may be several reasons for a failure/delay in convergence. One of them is the limited sampling of the parameter space, which is very likely a result of a too wide parameter range selection. Another reason may be high uncertainty in parameter–property correlation, which is likely due to the limited representation of the effects of these parameters in FF-training set. The increase in uncertainty is caused by inconsistent response in target property values to alteration of parameters in the first stage. This issue can be addressed by expanding FF-training set by adding reference property values that are correlated with the parameters of interest and less sensitive to change in parameter values. This expansion stabilizes the response to parameter alterations and reduces the uncertainty in target properties in ML-training set. Such high uncertainty is not only a problem for INDEEDopt, it may also affect the force field quality during production, because when the force field is trained specifically for very unstable reference properties, transferability of the force field to new environments that can be faced during production simulations is limited.
Flow diagram of INDEEDopt framework. Red boxes represent the three main stages of the framework: sampling with initial design algorithms, deep learning model training, and optimization using brute-force method. Blue diamonds represent the conditional transition points in the algorithm. Dark blue dashed boxes represent input files and settings given by the FF developer. The input files that are used in INDEEDopt are same as the ones used in conventional method. Dark blue box represents output files, which are the optimized force field parameters. The flow diagram of INDEEDopt is in alignment with on-the-fly learning schemes. Still, it is conceptually innovative due to smartly designing the parameter search and use these data to train a DL model, which is further used for optimization purposes.
A scatter plot of INDEEDopt predictions versus the ReaxFF calculations (i.e., true values). The blue dots are the data points and the red line is the linear trend line. The data points and the percent difference between the predictions and the true values can be seen in Supplementary Table 1.
The third stage of INDEEDopt is when the parameter optimization is performed by minimizing the Eq. (2).
where xi,ML and xi,REF are the target property obtained by trained DL model, and reference property values in FF-training set, respectively, and wi is the weight factor, which can be used to adjust the importance of property values during parametrization. Production of target property values using ReaxFF-MD code takes a significant amount of time during optimization, especially when the reference data set is extensive. This time issue limits the optimization quality as most of the developed algorithms tend to limit their parameter space sampling. To address this issue, INDEEDopt framework calculates the target properties using the trained DL model predictions instead of calling ReaxFF-MD code in the optimization stage. A well-trained DL model is capable of predicting target properties with reasonable accuracy in 3 ms, which is at least three orders of magnitude faster than using ReaxFF-MD code. Therefore, INDEEDopt is capable of exploring a wider parameter space more thoroughly. In addition, the ML-training set, which is generated to train DL model, has information about whole parameter space (in other words, it has the information for several local minima), and therefore can be used to train several parameters sets in parallel by targeting the best local minima in a smaller parameter range similar to ref. 28.
We performed optimization using two different approaches, namely brute-force method and Minimum Energy Design (MED) method, and compared the performances in the “Results” section. Additionally, because the optimization stage of INDEEDopt is separated from other stages, any other optimization algorithm can be integrated into the workflow. The brute-force optimization assigns randomly generated values to all parameters at once and calculates the discrepancy (y) between the target and reference property values. This process repeats until stopped by the force field developer when the discrepancy value is satisfactory. The criterion used to stop the optimization in this work was to produce a lower discrepancy than the one obtained by the conventional optimization method. However, if not assigned by the user, INDEEDopt uses the lowest discrepancy producing parameter set from the ML-training set. Even though the brute-force optimization is advantageous in terms of transitioning between local minima, it is not computationally efficient to be used in high-dimensional problems; therefore, we implemented the MED method, which can detect local minima faster as also demonstrated in ref. 38.
The MED is a smart active learning strategy that can be used to efficiently and adaptively explore the parameter space and identify combinations with low discrepancy between the target and reference property values. Proposed and developed by Joseph et al.39, it is typically used to explore large, complex spaces, quickly carving out “bad” (with large total discrepancies in our case), and producing more and more “good” combinations. A completely automated version of the algorithm based on ref. 40 is implemented in the R package mined. It requires the user to select a set of initial parameter combinations and a computer code (in our case the DL model) that calculates the total discrepancy between the target and reference property values for any given parameter combination. A more detailed description of the implementation of MED algorithm can be found in ref. 38.
Applications of INDEEDopt
The INDEEDopt framework was tested for two different force field development cases, namely, a Ni–Cr binary system for alloy applications and a W–S–C–O–H quinary system for 2D materials applications. The Ni–Cr system is relatively simple, due to the absence of angular and dihedral terms, which in turn requires optimization of a small number of parameters. The W–S–C–O–H system is more complex due to several reasons: (1) it has angular and dihedral interactions involved in the training set; (2) the number of parameters to be optimized and the size of the FF-training set is larger than the Ni–Cr one; (3) the force field training set includes both condensed and molecular phases together; therefore, W–S–C–O–H training was selected as a secondary test case to evaluate the performance of INDEEDopt framework. The same force fields were also parametrized using the conventional method, which is the most commonly used method in the ReaxFF literature24. All the settings related to energy minimization, molecular geometries in the FF-training set, and weights factors for discrepancy calculations were kept the same for the optimization using INDEEDopt and the conventional force field development method.
Ni–Cr force field optimization
The ReaxFF training set of the Ni–Cr alloy system is composed of 90 different reference property values, including equations of state and heat of formation energies for different Ni–Cr compositions in fcc and bcc phases. A total of 16 parameters were optimized, which involve bond-order terms, bond energy terms, over coordination terms, charge polarization terms, and Coulombic interaction terms. A detailed full list of parameters is given in Supplementary Table 2.
Using OMLHD algorithm, a total of 79,635 samples were created to be used as the ML-training set. Each parameter has a corresponding anticipated value range, mostly coming from the chemical nature of the element; however, some parameter ranges are determined by experience. The ranges used for each parameter (in other words, upper and lower limits of parameter search) are given in Supplementary Table 2. The determination of the ranges is important because wider ranges require more sampling, which is computationally inefficient, may also delay the optimization depending on the complexity of the problem. While exploring the parameter space, some parameter combinations produce meaningless inter- or intra-atomic interactions, and if the ranges are large, a significant amount of the sampling is conducted unnecessarily. For example, the Ni–Cr training set uses a very wide range for parameter-1, which is the bond energy term between Ni and Cr. Our analysis of the correlation between parameter-1 and remaining parameters (Supplementary Fig. 2) shows that a significant part (~80%) of the parameter range produces a large discrepancy meaning that the interatomic interactions are not usable for production simulations. INDEEDopt labels these large discrepancy regions as “infeasible” and remaining is referred to as “feasible”. After a first complete scan of the parameter space, INDEEDopt focuses on feasible regions and avoid sampling the infeasible parts of the parameter space, which avoids spending time to learn unnecessary parts of the parameter space.
A comparison of total discrepancy obtained by INDEEDopt and conventional method for different sections of the training set is given in Fig. 5. A detailed comparison of each FF-training set element can be seen in Supplementary Fig. 3. Most of the target properties in the training set are improved using INDEEDopt except some equation of states values Ni3Cr and Ni2Cr in FCC and BCC phases, respectively. A significant improvement is observed in the heat of formation energetics, and the lowest total error sum obtained using the conventional method was calculated as 5614.3, which was reduced to 2804.0 using INDEEDopt with brute force and 2316.2 using INDEEDopt with MED algorithm. During the optimization using the conventional method, the heat of formation and equations of state parts of the force field were inversely correlated; therefore, each part was optimized by compromising the accuracy of the other, and both parts were not able to be optimized at the same time. The reason why INDEEDopt succeeded is due to finding another local minimum in parameter space that can satisfy both parts of the force field. The significant differences between some of the final parameter values obtained by INDEEDopt and conventional method also show that the optimum parameter sets are detected in different regions of the parameter space (Fig. 6). The final parameter values obtained by both methods are found in Supplementary Table 2. Some of the parameters were optimized to similar values by both optimization methods, which may be due to a narrow parameter search range for these specific parameters assigned by the force field developer. INDEEDopt outperformed the conventional method in terms of timings by producing an optimized parameter set in around 2 days compared to 2 weeks by an experienced force field developer. However, we should note that the conventional method was run on a single core while INDEEDopt was run on a hundred cores.
The stacked bar graph representation of the difference between the parameter values in the nickel–chromium training set optimized using INDEEDopt with brute force and conventional method. The red represents the conventional, and yellow represents INDEEDopt version of the same parameter. The difference between the percentage values corresponds to the difference between predicted values by each method. The value 50% corresponds to a situation when both methods predict the same values. If the predicted parameter values are negative, then the percentage value is given as negative.
The overall improvement (i.e., reduction in total error) of the Ni–Cr force field translates into physical properties. The lattice expansion for Ni and Cr phases with the increase in temperature is calculated by INDEEDopt optimized force field, conventionally optimized force field, and compared with experimental values (Fig. 7). According to our analysis, the INDEEDopt optimized force field produces a superior result for Cr (BCC) phase compared to the conventionally optimized one, and it behaves similarly for Ni (FCC) phase. Additionally, energy–volume curves for each NinCrm phase can be seen in Supplementary Fig. 4.
W–S–C–O–H force field optimization
The parametrization of the W–S–C–O–H quinary system is more complex than the binary alloy case because the FF-training set involves both molecular and condensed phase parts. The presence of a condensed phase brings extra challenges to the optimization problem. One of the challenges is that the molecular and condensed phase may be best optimized separately in different local minima in the parameter space, meaning that a well-optimized molecular part does not necessarily produce low discrepancy for condensed phase part as well. In addition, the condensed phase part requires the usage of large molecular configurations to represent a crystalline phase. For example, the W–S–C–O–H training set has several geometries that are composed of hundreds of atoms, which makes these properties more sensitive to alteration in parameter values, and therefore brings considerable uncertainty to the parameter–property correlations. An additional complexity to the optimization problem comes from the size of the FF-training set. There are 289 reference properties in FF-training set, which is significantly higher than most data sets used in literature to test newly developed optimization procedures. The FF-training set includes heat of formation values for both molecular and condensed phase, defect and ad-atom energetics, some application-specific reaction energetics, strain energetics, and bond and angle energies. The total number of parameters to be optimized is 68, which include bond, angle, and off-diagonal terms. A detailed list of optimized parameters is given in Supplementary Table 3.
INDEEDopt produced a total of 44,700 samples until the DL model is trained to mimic ReaxFF-MD with reasonable accuracy. The total discrepancy sum for different parts in the FF-training set is given in Fig. 8 and compared with the ones calculated using the conventional method. A more detailed comparison is also given in Supplementary Fig. 5. A detailed explanation for each FF-training set part is given in Supplementary Note 2. The difference between the final optimized parameter values obtained by both methods can be seen in Fig. 9. As can be seen from the bar graphs, INDEEDopt produced a lower total error than the conventional method for angle scan, bond scan, and defect/ad-atom parts of the force field. The lowest total error sum obtained using the conventional method was calculated as 5314.4, which was reduced to as low as 5250.3 using INDEEDopt with brute force and 5705.5 using INDEEDopt with MED algorithm. The amount of time spent on optimization was approximately 2 months with the conventional method and was 5 days with INDEEDopt with brute-force algorithm. In addition, the usage of MED instead of the brute-force algorithm reduced this time to 4 days. Even though the MED algorithm was not able to produce the lowest error, the produced value is acceptable to be used in production simulations. We should note that the brute-force optimization criterion in this study was to do better than the conventional method; therefore, the optimization was stopped after the discrepancy value was below the one produced by the conventional method. However, the parameter set was very well developed by experienced FF developers using the conventional method; therefore, it was a challenging test. INDEEDopt spent approximately one day, around 30 million calculations, at the optimization stage to predict a lower discrepancy point than the conventionally obtained one. Therefore, it may not be guaranteed to predict a better point by running the optimization longer because: (1) INDEEDopt predicted a local minimum that is very different from the conventionally obtained one, which can be seen in Fig. 8, and there may not be a better point accessible by our optimization method in this specific low discrepancy region; (2) the DL model may not be able to make predictions as sensitive as ReaxFF-MD, which prevents revealing a better point. It may be possible to further improve the parametrization by assigning INDEEDopt-predicted parameters as a starting point to the conventional method by assigning a narrow parameter search range.
The total error values corresponding to different parts of the tungsten–sulfide–carbon–hydrogen FF-training set calculated by INDEEDopt with brute force and conventional methods. An explanation of the naming for different parts of FF-training set can be seen in Supplementary Note 2, and also same structures are used in ref. 6.
The stacked bar graph representation of the difference between the parameter values in the tungsten–sulfide–carbon–hydrogen training set optimized using INDEEDopt with brute force and conventional method. The red represents the conventional and yellow represents INDEEDopt version of the same parameter. The difference between the percentage values corresponds to the difference between predicted values by each method. The value 50% corresponds to a situation when both methods predict the same values. If the predicted parameter values are negative, then the percentage value is given as negative.
Discussion
An initial design enhanced DL-based optimization framework has been developed to parametrize ReaxFF force fields. The INDEEDopt framework was tested on two different training sets, and the performance was examined comparing with the conventional method. The tests were conducted on Ni–Cr and W–S–C–O–H training sets, of which lateral is the most challenging due to a large number of properties and diversity in the training set. The framework could produce a lower total discrepancy between ReaxFF values and reference property values compared to the conventional method. The time required for optimization calculations is also lower using INDEEDopt due to its ability to run on multiple processors. Another advantage of INDEEDopt framework is its ability to be used as an initial parameter generator that can be combined with other optimization methods, which can be valuable in avoiding local minima stucking problem in some other optimization methods and also to fine-tune force fields. Additionally, INDEEDopt can be used by any software that can run ReaxFF-MD energy minimization simulations, which makes it open to a wider user population.
This version of INDEEDopt is fully capable of providing a time-efficient tool for optimizing ReaxFF force fields with sufficient quality. However, there are some challenges that can be addressed to improve the framework further in future versions. The sampling scheme (first stage of the framework) used in INDEEDopt is computationally the most demanding part due to the high dimensionality of the problem, which can be addressed by developing problem-specific initial design methods. With the implementation of design of experiments-based optimization procedure for neural network hyperparameter tuning, desired accuracies from the DL model can be achieved in shorter periods of time. The future versions of INDEEDopt will also include subroutines that conduct statistical analysis to give feedback about the FF-training set, which will be very valuable to further improve FF quality. We believe that our framework provides major progress towards the process of ReaxFF parameterization. A Python 3 implementation of INDEEDopt framework is available at https://github.com/mertyigit/INDEEDopt for public use.
Methods
Computational details
INDEEDopt requires calculation of each reaction in ReaxFF training set to produce a data set, which is used to train a DL model. These calculations were performed as energy minimization and conducted at 0 K. The number of iterations were decided depending on the ReaxFF training set. The W–S–C–O–H training set involves condensed phase calculations, which require more iterations than molecular calculations; therefore, we used 1000 as the maximum number of iterations.
INDEEDopt is a parallel algorithm, which can perform ReaxFF simulations simultaneously. Calculations were performed on identical 20-core nodes (Intel Xeon Gold 6248 2.5G) using five in parallel. The GPU-based calculations were performed on identical NVIDIA Tesla P100s.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Code availability
A Python 3 implementation of INDEEDopt framework is available at https://github.com/mertyigit/INDEEDopt for public use.
References
Tersoff, J. New empirical approach for the structure and energy of covalent systems. Phys. Rev. B 37, 6991–7000 (1988).
Phillpot, S. R. et al. Charge Optimized Many Body (COMB) potentials for simulation of nuclear fuel and clad. Comput. Mater. Sci. 148, 231–241 (2018).
Brenner, D. W. et al. A second-generation reactive empirical bond order (REBO) potential energy expression for hydrocarbons. J. Phys. Condens. Matter 14, 783–802 (2002).
van Duin, A. C. T., Dasgupta, S., Lorant, F. & Goddard, W. A. ReaxFF: a reactive force field for hydrocarbons. J. Phys. Chem. A 105, 9396–9409 (2001).
Senftle, T. P. et al. The ReaxFF reactive force-field: development, applications and future directions. Npj Comput. Mater. 2, 15011–15011 (2016).
Ostadhossein, A. et al. ReaxFF reactive force-field study of molybdenum disulfide (MoS2). J. Phys. Chem. Lett. 8, 631–640 (2017).
Osti, N. C. et al. Influence of metal ions intercalation on the vibrational dynamics of water confined between MXene layers. Phys. Rev. Mater. 1, 65406–65414 (2017).
Sang, X. et al. In situ atomistic insight into the growth mechanisms of single layer 2D transition metal carbides. Nat. Commun. 9, 2266–2275 (2018).
Hasanian, M., Mortazavi, B., Ostadhossein, A., Rabczuk, T. & van Duin, A. C. T. Hydrogenation and defect formation control the strength and ductility of MoS2 nanosheets: reactive molecular dynamics simulation. Extreme Mech. Lett. 22, 157–164 (2018).
Sang, X. et al. Atomic defects and edge structure in single-layer Ti3C2Tx MXene. Microsc. Microanal. 23, 1704–1705 (2017).
Lotfi, R., Naguib, M., Yilmaz, D. E., Nanda, J. & van Duin, A. C. T. A comparative study on the oxidation of two-dimensional Ti3C2 MXene structures in different environments. J. Mater. Chem. A 6, 12733–12743 (2018).
Raju, M., van Duin, A. & Ihme, M. Phase transitions of ordered ice in graphene nanocapillaries and carbon nanotubes. Sci. Rep. 8, 3851–3862 (2018).
Achtyl, J. L. et al. Aqueous proton transfer across single-layer graphene. Nat. Commun. 6, 6539–6546 (2015).
Raju, M., Ganesh, P., Kent, P. R. & van Duin, A. C. Reactive force field study of Li/C systems for electrical energy storage. J. Chem. Theory Comput. 11, 2156–2166 (2015).
Merinov, B. V., Mueller, J. E., van Duin, A. C., An, Q. & Goddard, W. A. III ReaxFF reactive force-field modeling of the triple-phase boundary in a solid oxide fuel cell. J. Phys. Chem. Lett. 5, 4039–4043 (2014).
Zheng, Y. et al. Modeling and in situ probing of surface reactions in atomic layer deposition. ACS Appl. Mater. Interfaces 9, 15848–15856 (2017).
Liu, S., van Duin, A. C. T., van Duin, D. M., Liu, B. & Edgar, J. H. Atomistic insights into nucleation and formation of hexagonal boron nitride on nickel from first-principles-based reactive molecular dynamics simulations. ACS Nano 11, 3585–3596 (2017).
Zhang, C., van Duin, A. C. T., Seo, J. W. & Seveno, D. Weakening effect of nickel catalyst particles on the mechanical strength of the carbon nanotube/carbon fiber junction. Carbon 115, 589–599 (2017).
Ostadhossein, A., Yoon, K., van Duin, A. C. T., Seo, J. W. & Seveno, D. Do nickel and iron catalyst nanoparticles affect the mechanical strength of carbon nanotubes? Extreme Mech. Lett. 20, 29–37 (2018).
Shin, Y. K., Gai, L., Raman, S. & van Duin, A. C. Development of a ReaxFF reactive force field for the Pt-Ni alloy catalyst. J. Phys. Chem. A 120, 8044–8055 (2016).
Shin, Y. K., Kwak, H., Vasenkov, A. V., Sengupta, D. & van Duin, A. C. T. Development of a ReaxFF reactive force field for Fe/Cr/O/S and application to oxidation of butane over a pyrite-covered Cr2O3 catalyst. ACS Catal. 5, 7226–7236 (2015).
Russo, M. F. & van Duin, A. C. T. Atomistic-scale simulations of chemical reactions: bridging from quantum chemistry to engineering. Nucl. Instrum. Methods Phys. Res. B 269, 1549–1554 (2011).
Shchygol, G., Yakovlev, A., Trnka, T., van Duin, A. C. T. & Verstraelen, T. ReaxFF parameter optimization with Monte-Carlo and evolutionary algorithms: guidelines and insights. J. Chem. Theory Comput. 15, 6799–6812 (2019).
van Duin, A. C. T., Baas, J. M. A. & van de Graaf, B. Delft molecular mechanics: a new approach to hydrocarbon force fields. J. Chem. Soc. Faraday Trans. 90, 2881–2895 (1994).
Iype, E., Hutter, M., Jansen, A. P., Nedea, S. V. & Rindt, C. C. Parameterization of a reactive force field using a Monte Carlo algorithm. J. Comput. Chem. 34, 1143–1154 (2013).
Larsson, H. R., van Duin, A. C. & Hartke, B. Global optimization of parameters in the reactive force field ReaxFF for SiOH. J. Comput. Chem. 34, 2178–2189 (2013).
Dittner, M., Muller, J., Aktulga, H. M. & Hartke, B. Efficient global optimization of reactive force-field parameters. J. Comput. Chem. 36, 1550–1561 (2015).
Jaramillo-Botero, A., Naserifar, S. & Goddard, W. A. 3rd General multiobjective force field optimization framework, with application to reactive force fields for silicon carbide. J. Chem. Theory Comput. 10, 1426–1439 (2014).
Rice, B. M., Larentzos, J. P., Byrd, E. F. & Weingarten, N. S. Parameterizing complex reactive force fields using multiple objective evolutionary strategies (MOES): Part 2: transferability of ReaxFF models to C-H-N-O energetic materials. J. Chem. Theory Comput. 11, 392–405 (2015).
Larentzos, J. P., Rice, B. M., Byrd, E. F., Weingarten, N. S. & Lill, J. V. Parameterizing complex reactive force fields using multiple objective evolutionary strategies (MOES). Part 1: ReaxFF models for cyclotrimethylene trinitramine (RDX) and 1,1-diamino-2,2-dinitroethene (FOX-7). J. Chem. Theory Comput. 11, 381–391 (2015).
Guo, F. et al. Intelligent-ReaxFF: evaluating the reactive force field parameters with machine learning. Comput. Mater. Sci. 172, 109393–109404 (2020).
Furman, D., Carmeli, B., Zeiri, Y. & Kosloff, R. Enhanced particle swarm optimization algorithm: efficient training of ReaxFF reactive force fields. J. Chem. Theory Comput. 14, 3100–3112 (2018).
Joseph, V. R. & Hung, Y. Orthogonal-maximin Latin hypercube designs. Stat. Sin. 18, 171–186 (2008).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Glorot, X., Bordes, A. & Bengio, Y. Deep sparse rectifier neural networks. JMLR Workshop Conf. Proc. 15, 315–323 (2011).
Breuel, T. M. The effects of hyperparameters on SGD training of neural networks. Preprint at https://arxiv.org/abs/1508.02788 (2015).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2015).
Song, Y. et al. CLAIMED: A CLAssification-Incorporated Minimum Energy Design to explore a multivariate response surface with feasibility constraints. Preprint at https://arxiv.org/abs/2006.05021 (2020).
Joseph, V. R., Dasgupta, T., Tuo, R. & Wu, C. F. J. Sequential exploration of complex surfaces using minimum energy designs. Technometrics 57, 64–74 (2015).
Joseph, V. R., Wang, D., Gu, L., Lv, S. & Tuo, R. Deterministic sampling of expensive posteriors using minimum energy designs. Technometrics 61, 297–308 (2019).
Acknowledgements
The authors acknowledge partial funding support from U.S. National Science Foundation under Award No. DMR-1842922, DMR-1842952, DMR-1539916, and MRI-1626251.
Author information
Authors and Affiliations
Contributions
M.Y.S., T.D., Y.H., and A.C.T.v.D. came up with the procedure idea. The code is written by M.Y.S. The manuscript was written by M.Y.S. through contributions of T.D., Y.H., A.C.T.v.D. The ReaxFF training sets are provided by N.N., Y.G., and A.C.T.v.D. The minimum energy design method is implemented by Y.S. All authors have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sengul, M.Y., Song, Y., Nayir, N. et al. INDEEDopt: a deep learning-based ReaxFF parameterization framework. npj Comput Mater 7, 68 (2021). https://doi.org/10.1038/s41524-021-00534-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-021-00534-4
This article is cited by
-
Multiscale modelling of transport in polymer-based reverse-osmosis/nanofiltration membranes: present and future
Discover Nano (2024)
-
Mixing ReaxFF parameters for transition metal oxides using force-matching method
Journal of Molecular Modeling (2022)