Abstract
We compared cell-free DNA (cfDNA) results at MBC diagnosis in patients who developed brain metastases (BM) vs those without (non-BM) to understand genomic predictors of BM. Patients with cfDNA testing at MBC diagnosis (Guardant360®, 73 gene next generation sequencing) were identified. Clinical and genomic features of BM and non-BM were compared (Pearson’s/Wilcoxon rank sum tests). Eighteen of 86 patients (21%) with cfDNA at MBC diagnosis developed BM. Comparing BM vs non-BM, a higher prevalence of BRCA2 (22% vs 4.4%, p = 0.01), APC (11% vs 0%, p = 0.005), CDKN2A (11% vs 1.5%, p = 0.05), and SMAD4 (11% vs 1.5%, p = 0.05) was observed. Seven of 18 BM had ≥1 of the following 4 mutations in baseline cfDNA: APC, BRCA2, CDKN2A or SMAD4 vs 5/68 non-BM (p = 0.001). Absence of this genomic pattern had a high negative predictive value (85%) and specificity (93%) in excluding BM development. Baseline genomic profile varies in MBC that develops BM.
Similar content being viewed by others
Introduction
Metastatic breast cancer (MBC) can vary in disease presentation, subtype, and molecular profiles, and these variable features may profoundly impact patient outcomes1,2,3. Brain metastases in patients with MBC cause significant morbidity and mortality4, and may affect a proportion of patients with breast cancer5,6. Some predictors for the development of brain metastases such as disease subtype, presence of lung metastases, and extensive visceral involvement are well-documented7. Prior studies have demonstrated that genomic profiles may vary in breast cancer, contributing to disease presentation and outcomes8,9,10,11,12,13. However, specific genomic predictors of brain metastases in patients with MBC are not well-understood, given that brain tumor tissue is not routinely obtained for genotyping due to the difficulty of obtaining brain tissue via surgical procedures. In recent years, cell-free DNA (cfDNA) has emerged as a strategy to identify tumor mutations of therapeutic and prognostic significance14,15,16,17,18, and offers the advantage of being much less invasive than a tumor tissue biopsy. It is not known whether cfDNA could potentially be used to help identify patients with MBC who may be at higher risk for the development of brain metastases. Understanding potential genomic risk factors for the development of brain metastases could help provide rationale for studying screening and/or therapeutic interventions to reduce the burden of brain metastases in genomically high-risk populations.
The purpose of this study was to explore the potential utility of cfDNA for the identification of patients with MBC with a high risk of developing brain metastases. We compared tumor genotyping results via cfDNA collected at MBC diagnosis in patients who developed brain metastases after MBC diagnosis (BM) with those who did not develop brain metastases (non-BM) to understand potential differences in baseline genomic characteristics.
Results
Clinical characteristics of BM vs non-BM
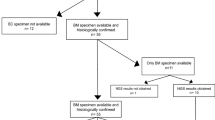
Of 86 patients with MBC who had cfDNA collected at the time of MBC diagnosis, 18 (21%) developed brain metastases during their disease course, of which 5 patients had brain metastases at the time of MBC diagnosis (Fig. 1). Table 1 depicts the baseline characteristics of both cohorts. The median time to development of brain metastases after cfDNA testing was 11.5 months (range 0–36 months). The median follow-up period was similar for both cohorts (BM: median 19.1 months, range 6.1–120.4 months; non-BM: median 27.2 months, range 6.2–53.4 months). Patients with BM vs non-BM had a similar distribution of disease subtype, visceral vs non-visceral disease, de-novo vs recurrent MBC, extracranial sites of disease, distribution of ethnicity (majority white women), and age at MBC diagnosis. Notably, there was a similar disease subtype distribution in BM (HR + /HER2:67%, HER2 + :11%, TNBC:17%) and non-BM (HR + /HER2-:69%, HER2 + :12%, TNBC:19%). The majority of patients with and without brain metastases were previously untreated for MBC at the time of cfDNA collection. Patients with brain metastases were treated with surgery alone (22%), surgery and radiation (5.5%), radiation alone (16%), radiation and systemic therapy (22%), systemic therapy alone (22%), and palliative care (11%). Median survival after the development of brain metastases was 11.6 months. Median overall survival after MBC diagnosis for BM was 26.4 months compared to 54.6 months for non-BM.
Genomic characteristics of BM vs non-BM
Of BM patients, 94% (17) had ≥1 detectable cfDNA mutation, while 91% (62) of non-BM patients had ≥1 detectable cfDNA mutation (p = 0.65). Patients with BM had a median of 4.5 mutations in baseline cfDNA compared to patients with non-BM who had a median of 3 (p = 0.31). Median number of mutations also did not vary significantly when corrected for disease subtype, possibly due to the small sample size (hormone receptor positive (HR+): BM median 2 mutations, non-BM 2; HER2+:BM 3, non-BM 2; triple negative breast cancer (TNBC): BM 3, non-BM 1). Patients with BM also had a median maximum mutant allele fraction (MAF) of 4.1% compared to 3.2% in the non-BM cohort.
The mutation pattern varied between cohorts as depicted in Fig. 2. Notably, among patients who developed BM compared to non-BM, higher prevalence of BRCA2 mutations (cases: 4/18 vs 3/68; percentage: 22.2% vs 4.4%, p = 0.01), APC mutations (cases: 2/18 vs 0/68; percentage: 11.1% vs 0.0%, p = 0.005), SMAD4 mutations (cases: 2/18 vs 1/68; percentage: 11.1% vs 1.5%, p = 0.05) and CDKN2A mutations (cases 2/18 vs 1/68; percentage: 11.1% vs 1.5%, p = 0.05) were observed. Supplementary Table 1 compares the prevalence of individual mutations between BM and non-BM. A higher prevalence of BRCA1 was seen in BM vs non-BM (11.1% vs 2.9%), although this difference is statistically nonsignificant (p = 0.14). Of the BRCA1/2 mutations in this cohort, only 1 patient had a known germline BRCA2 mutation (in BM cohort). Supplementary Table 2 classifies the mutations identified in the BM cohort as pathogenic, uncertain significance, or synonymous variants. Supplementary Fig. 1 compares frequencies of amplified genes between BM and non-BM; no statistically significant differences were seen between these cohorts.
Supplementary Fig. 2 depicts the mutation spectrum observed in 5 patients with brain metastases at the time of cfDNA testing at MBC diagnosis; notably, BRCA1 (20%), BRCA2 (20%), CDKN2A (40%), and APC (40%) were seen in cfDNA in this cohort.
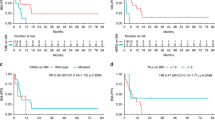
The mutation spectrum for individual patients with BM or non-BM was then plotted, as depicted in Figs. 3, 4, to evaluate patterns of expression among patients with BM (Fig. 3) vs non-BM (Fig. 4). We were particularly interested in the expression of APC, BRCA2, CKDN2A, and SMAD4 given the statistically significant higher prevalence seen in BM within the entire cohort. We observed that 7/18 patients who developed BM had at least 1 of the following 4 mutations present in baseline cfDNA: APC, BRCA2, CDKN2A, or SMAD4 compared to only 5/68 patients with non-BM who had at least 1 of those 4 mutations (p = 0.001). Thus, this genomic pattern (having at least 1 of the following 4 mutations: APC, BRCA2, CDKN2A, or SMAD4) has a positive predictive value of 58%, negative predictive value of 85%, sensitivity of 39%, and specificity of 93% for prediction of early onset brain metastases in patients with MBC.
For patients with brain metastases, 5/18 (28%) underwent surgery or a brain biopsy. Due to the retrospective nature of this analysis, while we did not have brain tumor samples from all patients, four patients had tumor genotyping results available from the brain tumor specimens, with 3 of the cases demonstrating a concordant finding in cfDNA (at MBC diagnosis) and brain tumor tissue, as indicated in Supplementary Fig. 3. In the last case, the majority of mutations found on brain tumor tissue genotyping were not covered by the cfDNA panel sent at the time.
Discussion
Brain metastases are associated with significant morbidity and mortality from MBC. A better understanding of risk factors for the development of brain metastases may enable the earlier identification and/or treatment of at-risk patients. While some features of breast tumors that predispose to the development of brain metastases are known, such as disease subtype, genomic predictors of brain metastases in patients with MBC are not well-delineated, given the inherent challenge of obtaining brain tumor tissue for genotyping. CfDNA can identify oncogenic mutations from DNA shed from tumor cells14,15,16,17,18. In this study, we compared cfDNA results collected at the time of MBC diagnosis in patients who developed brain metastases at or following MBC diagnosis with those who did not develop brain metastases.
Our results suggest that patients with MBC who subsequently develop early brain metastases may have different baseline cfDNA genomics. We observed a significantly higher prevalence of APC, BRCA2, CDKN2A and SMAD4 mutations in patients who developed brain metastases than those without brain metastases.
The presence of differences in the genomic landscape of patients with MBC who go on to develop brain metastases is intriguing. As our median follow up period in both cohorts was about 2 years, the genomic differences we have observed may help to identify patients with MBC who may be at risk for the early development of brain metastases. Our finding of a higher prevalence of brain metastases in patients with cfDNA APC, BRCA2, CDKN2A or SMAD4 mutations is intriguing, and merits further investigation in a larger prospective analysis controlling for disease subtype and presence of extracranial visceral disease. Interestingly, mutation burden was similar for BM vs non-BM in cfDNA, as was the presence of extracranial visceral disease, age at time of MBC diagnosis, and disease subtype in our cohort. Prior literature has suggested that germline BRCA1/2 mutations may be associated with the development of brain metastases19,20,21. A study noted that CNS metastases frequently occurred in both BRCA1 and BRCA2 germline carriers (53% BRCA1 and 50% BRCA2 carriers), but only the BRCA2 mutation had a statistically significant association with brain metastasis on multivariable analysis20. A second cohort also identified a high rate of brain metastases in patients with germline BRCA1/2 mutations22. Although only 1 patient in our BM cohort was a known germline BRCA2 carrier, it is possible that somatic BRCA2 mutations may exhibit similar clinical behavior as germline BRCA2 mutations23. Another study has demonstrated that brain metastases often demonstrate loss of the APC gene24. In addition, CDKN2A was found to be a commonly mutated gene in brain metastases from lung adenocarcinoma25, and a higher prevalence of CDKN2A/p16 has also been observed in the lymph nodes of patients with breast cancer who developed brain metastases26. In lung adenocarcinoma, SMAD4 has been detected in cerebrospinal fluid circulating tumor DNA27. SMAD4 has also been implicated in the pathogenesis of breast cancer28.
Further prospective research is needed to validate the genomic pattern we have identified (i.e. presence of at least one of the 4 genes: APC, BRCA2, CDKN2A and SMAD4) that was more commonly observed in patients who developed brain metastases versus patients who did not experience brain metastases (7/18 vs 5/68, p = 0.001). Given the high specificity (93%) and negative predictive value (85%) of this genomic pattern in our cohort, this genomic pattern in cfDNA has the potential to serve as a genomic marker to identify patients with MBC who may have a lower risk of developing early brain metastases. As noted above, literature from other authors also lends support to the inclusion of APC, BRCA2, CDKN2A, and SMAD4. Indeed, this genomic pattern was identified in 3/5 patients with brain metastases at the time of MBC diagnosis in our cohort.
Future development of a preclinical model such as a cell line from a patient with MBC mutated with the genomic signature we identified could be used as a proof-of-concept of our hypothesis. Given the retrospective nature of our work, we were not able to conduct such an experiment in this study.
While the evaluation of cfDNA in patients with brain metastases is limited, prior studies have analyzed median levels of cfDNA in patients with glioblastoma or stage IV adenocarcinoma, demonstrating increases in cfDNA prior to tumor recurrence29. Other authors have also identified the presence of cfDNA mutations in patients with primary brain tumors30. Another approach that has been taken is to evaluate genomics using the CSF of patients with brain metastases31,32,33,34,35. The correlation of cfDNA and CSF findings is not well-established, and a potential direction for future research, since cfDNA offers the advantage of being a less invasive procedure. Research is also needed to help understand the correlation between cfDNA and brain tumor tissue mutation profiling; given our limited brain tissue sampling data, we were not able to study this correlation in our cohort, although concordance of genomic findings between baseline cfDNA at MBC diagnosis and brain tumor tissue genotyping was noted for 3 patients.
Additional limitations of our work include the retrospective nature of these analyses and modest sample size at a single institution limiting further subset analyses. Furthermore, as we used the Guardant360® platform, we are limited to the genotyping panel included in this assay. Using whole exome sequencing could potentially identify additional mutations of potential significance.
Our finding that the mutation spectrum of cfDNA collected at the time of MBC diagnosis may vary in patients who subsequently develop early brain metastases warrants further investigation. The prospective validation of genomic predictors using cfDNA could potentially guide precision medicine interventions to decrease the risk of developing brain metastases and/or support enhanced screening measures to enable earlier identification of brain metastases in high-risk populations.
Methods
This study was performed in compliance with the Declaration of Helsinki. The retrospective analyses were performed with IRB approval from an institutional protocol. Per IRB regulations, individual patient consent was not required for this retrospective analysis, although all patients had been consented for Guardant360® cfDNA testing prior to collection.
cfDNA
The cfDNA analysis was conducted as part of routine clinical practice via the Guardant360® assay (Guardant Health Inc., Palo Alto, CA). cfDNA was obtained from whole blood, with blood draw, shipment, plasma isolation, and cfDNA extraction procedures previously described36. Guardant360® is CLIA-certified, College of American Pathologists-accredited, New York State Department of Health-approved cfDNA next-generation sequencing (NGS) assay with analytical and clinical validation previously reported17,36,37. At the time of the study, Guardant360® included analysis of single nucleotide variants (SNVs) in 73- to 74-genes (the assay evolved over the course of the study period), as well as small insertions/deletion (indels), copy number amplifications, and gene rearrangement/fusions in a subset of genes. The reportable range for SNVs, indels, fusions, and CNAs is ≥0.04%, ≥0.02%, ≥0.04%, and ≥2.12 copies, respectively, with >99.9999% per-position analytic specificity36.
Comparison of baseline cfDNA genomics (at time of MBC diagnosis) in patients with MBC who developed brain metastases (BM) vs those without brain metastases (non-BM)
Patient population
In this study, we identified patients with MBC at the Massachusetts General Hospital who underwent cfDNA testing (detailed above) as part of routine clinical care at the time of MBC diagnosis between 1/2016–10/2019 with greater than or equal to 6 months of follow-up at our institution post-testing. From this cohort of patients, the subset of patients who developed brain metastases either at the time of or after cfDNA testing was identified. A retrospective review of medical records and pathology reports was conducted to determine demographics and tumor subtype. In addition, a retrospective review of Guardant360® reports was conducted to analyze the cfDNA for the number and type of mutations, as well as the maximum mutant allele fraction (MAF). For patients for whom brain tumor tissue was available, a comparison of the cfDNA results at MBC diagnosis with the brain tumor genotyping results was made.
Analysis
Clinical and genomic features of patients who developed brain metastases (BM) at or after cfDNA collection at MBC diagnosis, and those without brain metastases (non-BM) were compared using Pearson’s and Wilcoxon rank sum tests, with p ≤ 0.05 for statistical significance. Fisher’s exact test with p ≤ 0.05 for statistical significance was used to compare gene amplifications between BM vs non-BM.
A comparison of the mutational spectrum in patients who developed brain metastases versus those who did not was performed to help identify a panel of genes that might risk stratify patients for the development of brain metastases.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data used in this study is not publically available as it contains patient information including genomic/genetic information, which cannot be released as this is not covered by the informed consent signed by patients. Clarifications on the study data may be requested in writing to the corresponding author.
References
Kennecke, H. et al. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol.: Off. J. Am. Soc. Clin. Oncol. 28, 3271–3277 (2010).
Xiao, W. et al. Breast cancer subtypes and the risk of distant metastasis at initial diagnosis: a population-based study. Cancer Manag. Res. 10, 5329–5338 (2018).
Guo, Y. et al. Different breast cancer subtypes show different metastatic patterns: a study from a large public database. Asian Pac. J Cancer Prevent.: APJCP 21, 3587–3593 (2020).
Taillibert, S. & Le Rhun, É. Epidemiology of brain metastases. Cancer Radiother. 19, 3–9 (2015).
Arvold, N. D. et al. Brain metastases after breast-conserving therapy and systemic therapy: incidence and characteristics by biologic subtype. Breast Cancer Res. Treat. 136, 153–160 (2012).
Barnholtz-Sloan, J. S. et al. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J. Clin. Oncol.: Off. J Am. Soc. Clin. Oncol. 22, 2865–2872 (2004).
Brosnan, E. M. & Anders, C. K. Understanding patterns of brain metastasis in breast cancer and designing rational therapeutic strategies. Ann. Transl. Med. 6, 163 (2018).
Callens, C. et al. Molecular features of untreated breast cancer and initial metastatic event inform clinical decision-making and predict outcome: long-term results of ESOPE, a single-arm prospective multicenter study. Genome Med. 13, 44 (2021).
Bertucci, F. et al. Genomic characterization of metastatic breast cancers. Nature 569, 560–564 (2019).
Chan, J. J., Tan, T. J. Y. & Dent, R. A. Are there any clinically relevant subgroups of triple-negative breast cancer in 2018? J. Oncol. Pract. 14, 281–289 (2018).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 121, 2750–2767 (2011).
Liu, M. C. et al. PAM50 gene signatures and breast cancer prognosis with adjuvant anthracycline- and taxane-based chemotherapy: correlative analysis of C9741 Alliance. npj Breast Cancer 2, 15023 (2016).
Li, A., Schleicher, S. M., Andre, F. & Mitri, Z. I. Genomic Alteration in Metastatic Breast Cancer and Its Treatment. Am. Soc. Clin. Oncol. Educ. Book, 30–43, https://doi.org/10.1200/EDBK_280463 (2020).
Zheng, Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 20, 1479–1484 (2014).
Haber, D. A. & Velculescu, V. E. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 4, 650–661 (2014).
Aggarwal, C. et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 5, 173–180 (2019).
Lanman, R. B. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PloS One 10, e0140712 (2015).
Leighl, N. B. et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res.: Off. J Am. Assoc. Cancer Res. 25, 4691–4700 (2019).
Albiges, L. et al. Spectrum of breast cancer metastasis in BRCA1 mutation carriers: highly increased incidence of brain metastases. Ann. Oncol.: Off. J Eur. Soc. Med. Oncol. 16, 1846–1847 (2005).
Song, Y. et al. Patterns of recurrence and metastasis in BRCA1/BRCA2-associated breast cancers. Cancer 126, 271–280 (2020).
Ratner, E. et al. Increased risk of brain metastases in ovarian cancer patients with BRCA mutations. Gynecolog. Oncol. 153, 568–573 (2019).
Garber H. et al. The incidence and impact of brain metastases in patients with hereditary BRCA1/2 mutated invasive breast cancer in a prospectively followed cohort. J. Clin. Oncol. 38, 1096–1096.
Vidula, N. et al. Identification of somatically acquired BRCA1/2 mutations by cfDNA analysis in patients with metastatic breast cancer. Clin. Cancer Res.: Off. J Am. Assoc. Cancer Res. 26, 4852–4862 (2020).
Pećina-Šlaus, N., Nikuševa Martić, T., Zeljko, M. & Bulat, S. Brain metastases exhibit gross deletions of the APC gene. Brain Tumor Pathol. 28, 223–228 (2011).
Preusser, M. et al. Spectrum of gene mutations detected by next generation exome sequencing in brain metastases of lung adenocarcinoma. Eur. J. Cancer 51, 1803–1811 (2015).
Furet, E. et al. Increased risk of brain metastases in women with breast cancer and p16 expression in metastatic lymph-nodes. Oncotarget 8, 37332–37341 (2017).
Ma, C. et al. Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thoracic Cancer 11, 588–593 (2020).
Zhong, D. et al. Homozygous deletion of SMAD4 in breast cancer cell lines and invasive ductal carcinomas. Cancer Biol. Ther. 5, 601–607 (2006).
Faria, G., Silva, E., Da Fonseca, C. & Quirico-Santos, T. Circulating cell-free DNA as a prognostic and molecular marker for patients with brain tumors under perillyl alcohol-based therapy. Int. J. Mol. Sci. 19, https://doi.org/10.3390/ijms19061610 (2018).
Piccioni, D. E. et al. Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol. 8, Cns34 (2019).
Seoane, J., De Mattos-Arruda, L., Le Rhun, E., Bardelli, A. & Weller, M. Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann. Oncol.: Off. J Eur. Soc. Med. Oncol. 30, 211–218 (2019).
Shi, L. et al. P2.01-86 Genetic profiling of circulating cell-free DNA from cerebrospinal fluid and plasma in ALK-positive lung cancer with brain metastases. J. Thoracic Oncol. 13, S697–S698 (2018).
Pentsova, E. I. et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J. Clin. Oncol.: Off. J. Am. Soc. Clin. Oncol. 34, 2404–2415 (2016).
Pan, W., Gu, W., Nagpal, S., Gephart, M. H. & Quake, S. R. Brain tumor mutations detected in cerebral spinal fluid. Clin. Chem. 61, 514–522 (2015).
Wang, J. et al. Detection of tumor-derived DNA mutations in cerebrospinal fluid of patients with primary or metastatic brain tumors. J. Clin. Oncol. 35, 2070–2070 (2017).
Odegaard, J. I. et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 24, 3539–3549 (2018).
Zill, O. A. et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 24, 3528–3538 (2018).
Acknowledgements
We would like to acknowledge Ms. Caroline Weipert at Guardant Health for her clarifications on the Guardant360® methodology and assistance with analyzing gene amplifications. No research support was received for this study.
Prior presentation: This study was presented in part at the ASCO 2020 annual meeting (Vidula N et al. Comparison of the cell-free DNA genomics in patients with metastatic breast cancer (MBC) who develop brain metastases versus those without brain metastases. JCO. May 2020. 38(15_suppl): 1094–1094.).
Author information
Authors and Affiliations
Contributions
N.V. and A.B. designed the study. N.V. wrote the first draft of the manuscript. N.V., A.N., K.H., L.R., B.M., S.I., L.E., D.J., and A.B. all participated in data collection, interpretation, analysis, and review and approval of the final manuscript.
Corresponding author
Ethics declarations
Competing interests
No funding was provided for this study. Individual disclosures for co-authors are as noted below: A.N.: The author declares no competing financial and non-financial interests. K.S.: The author declares no competing financial and non-financial interests. L.R.: The author declares no competing financial and non-financial interests. B.M.: The author declares no competing financial and non-financial interests. S.I.: The author declares no competing financial and non-financial interests. L.E.: The author declares no competing financial and non-financial interests. N.V.: The author declares no competing non-financial interests but the following competing financial interests: Research funding (for clinical trials not related to this work) to the institution (Massachusetts General Hospital): Merck, Daehwa, Pfizer, Radius, and Novartis; Advisory board participation (single session, not related to this work): AbbVie, OncoSec, TerSera, and Gilead DJ: The author declares no competing non-financial interests but the following competing financial interests: Consulting fees from Novartis, Genentech, Syros, Eisai, Vibliome, Mapkure, and Relay Therapeutics. Contracted research with Novartis, Genentech, Syros, Pfizer, Eisai, Takeda, Ribon Therapeutics, Infinity, InventisBio, Cyteir, Blueprint Medicines and Arvinas. Ownership interest in Relay Therapeutics and PIC Therapeutics. A.B.: The author declares no competing non-financial interests but the following competing financial interests: Consultant/Advisory board: Pfizer, Novartis, Genentech, Merck, Radius Health, Immunomedics/Gilead, Sanofi, Daiichi Pharma/Astra Zeneca, Phillips, Eli Lilly, Foundation Medicine. Contracted Research/Grant (to institution): Genentech, Novartis, Pfizer, Merck, Sanofi, Radius Health, Immunomedics/Gilead, Daiichi Pharma/Astra Zeneca, Eli Lilly.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vidula, N., Niemierko, A., Hesler, K. et al. Utilizing cell-free DNA to predict risk of developing brain metastases in patients with metastatic breast cancer. npj Breast Cancer 9, 29 (2023). https://doi.org/10.1038/s41523-023-00528-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-023-00528-z