Abstract
The identification and molecular characterization of cellular hierarchies in complex tissues is key to understanding both normal cellular homeostasis and tumorigenesis. The mammary epithelium is a heterogeneous tissue consisting of two main cellular compartments, an outer basal layer containing myoepithelial cells and an inner luminal layer consisting of estrogen receptor-negative (ER−) ductal cells and secretory alveolar cells (in the fully functional differentiated tissue) and hormone-responsive estrogen receptor-positive (ER+) cells. Recent publications have used single-cell RNA-sequencing (scRNA-seq) analysis to decipher epithelial cell differentiation hierarchies in human and murine mammary glands, and reported the identification of new cell types and states based on the expression of the luminal progenitor cell marker KIT (c-Kit). These studies allow for comprehensive and unbiased analysis of the different cell types that constitute a heterogeneous tissue. Here we discuss scRNA-seq studies in the context of previous research in which mammary epithelial cell populations were molecularly and functionally characterized, and identified c-Kit+ progenitors and cell states analogous to those reported in the recent scRNA-seq studies.
Similar content being viewed by others
Previous studies to elucidate the cellular identities of mammary epithelial subpopulations have involved functional and molecular characterization by flow cytometric and functional (down to single cell) transplantation assays1,2,3,4,5,6,7,8,9,10,11,12,13,14, as well as, more recently, lineage-tracing studies15,16,17,18,19,20,21,22,23,24,25,26. Transplantation experiments have generally supported a model in which facultative MaSCs, cells capable of regenerating the epithelium when injected into a cleared mammary fat pad (one free of endogenous epithelium)1,27, are localized to the basal cell layer2,5,9,28,29,30. Progenitor cells, which are functionally defined by high colony-forming and proliferative potential in vitro and limited repopulating ability when transplanted into cleared fat pads, are localized to the luminal layer6,10,28,29. Differentiated cells do not transplant or generate colonies in vitro. The molecular profiling of mammary epithelial subpopulations functionally defined by their transplantation potential has been extensive9,17,31,32,33,34,35,36,37,38,39,40.
Supporting this model, in situ evidence, including lineage-tracing studies from early mammary development, puberty, and alveolargenesis during pregnancy, has shown that basal cells can contribute to the luminal layer19,41,42,43. We previously proposed, based on in situ analysis, that basal MaSCs located in the cap cell layer of terminal end buds (TEBS), the outermost cell layer of the specialized growth structure that drives ductal growth during puberty, are bipotent and produce daughter cells that contribute to both the basal and luminal cell lineages43. Lineage-tracing experiments from Rios et al.16 and Wang et al.15 were in agreement with transplantation data and our in situ analysis, suggesting that MaSCs in the developing postnatal gland are bipotent15,16,43. However, more recently, it has been shown that, rather than a transcriptionally defined bipotent TEB MaSC, a group of transcriptionally heterogeneous lineage-committed MaSCs mediate development of the pubertal mammary gland and contribute transiently to ductal expansion23, mirroring the organization and neutral drift of adult stem cells observed in the intestine44,45. This model of postnatal mammary gland development is in agreement with saturation, single-cell genetic, and neutral lineage-tracing studies demonstrating that bipotent fetal MaSCs (fMaSCs), first functionally and molecularly characterized (including single-cell gene expression analysis demonstrating molecular heterogeneity) by Spike et al.37, exist in the embryo, but that in the postnatal gland, basal and luminal lineages are maintained by separate lineage-committed stem/progenitor populations18,19,20,21,22,23,24,42,46,47,48. During oncogenic transformation, basal and luminal cell populations may lose this restricted lineage potential and acquire multipotency20,24,49,50.
Recent studies have used scRNA-seq, which unlike functional and population-based sequencing studies, allows for unbiased analysis of individual cells in a heterogeneous tissue, to decipher lineage hierarchies and cell states in the mammary epithelium51,52,53,54. To investigate cellular heterogeneity and lineage relationships in the human breast, Nguyen et al.51 performed scRNA-seq analysis on fluorescence-activated cell-sorted (FACS) breast epithelial cells and reported the identification of additional cell types within the three main mammary epithelial cell populations, previously identified as basal (B: CD49fHigh EPCAM+, K14+), luminal progenitors (L1: CD49f+ EPCAM+, ER−, K8/18+), and mature luminal (L2: CD49f− EPCAM+, ER+, K8/18+) cells8,10,51. Significantly, the authors detected replicating KIT+ cells in all three main populations (Basal, L1, and L2), suggesting that each cluster may be maintained by its own KIT+ progenitor cell population, and proposed a continuous lineage hierarchy connecting the basal lineage to the two luminal branches via a bipotent MaSC. Furthermore, the authors highlight adult luminal cells that co-express both luminal (KRT8/18) and basal (KRT14) markers in situ.
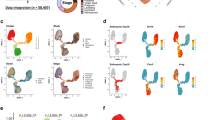
The receptor tyrosine kinase KIT (c-Kit) has previously been identified as a defining marker of mammary epithelial progenitor cells (summarized in Table 1) and of the cells of origin of BRCA1-mutation breast cancer, luminal ER− cells17,28,34,40,50,55. Similar to Nguyen et al.51, in Regan et al.28, we identified in the mouse, and also functionally tested via in vitro colony-forming assays and cleared mammary fat pad transplantation, c-Kit− and c-Kit+ cell states within each of the mammary epithelial basal (CD24+/Low Sca-1− CD49f+/High c-Kit− and c-Kit+), myoepithelial (CD24+/Low Sca-1− CD49f+/Low c-Kit− and c-kit+), luminal ER− (CD24+/High Sca-1− c-Kit+/Low and c-Kit+/High), and luminal ER+ (CD24+/Low Sca-1− c-Kit−, CD24+/Low Sca-1+ c-kit− and c-kit+) cellular compartments28. The expression of KIT, as well as the luminal markers KRT8/18 and ESR1 and basal marker KRT14, in each of Nguyen et al.’s human breast populations of B, Myo, L1.1, L1.2, and L2, are consistent with the expression levels reported in Regan et al.28 in the corresponding murine basal, myoepithelial, luminal ER– c-Kit+/High, luminal ER− c-Kit+/Low, and luminal ER+ cells, respectively (Fig. 1). The KIT+ cells identified by Nguyen et al.51 are therefore likely equivalent to the c-Kit+ progenitor cells previously reported in Regan et al.28, which was the first study to functionally characterize c-Kit as a progenitor marker in the mammary gland (Table 1). When discussing KIT as a progenitor cell marker, Nguyen et al. incorrectly cite Stingl et al.56 and Shehata et al.10. These papers, respectively, did not investigate or functionally test c-Kit as a progenitor marker in the mammary gland.
Nguyen et al.51 violin plots showing the expression pattern of progenitor marker KIT (a LHS), luminal genes ESR1 and KRT8 (b, c LHS), and basal gene KRT14 (d LHS) grouped by final cluster determination in human mammary epithelium. B = basal (containing facultative MaSCs), Myo = myoepithelial. Regan et al.28 gene expression in the different cellular subpopulations as determined by qPCR for progenitor gene c-Kit (a RHS) relative to comparator luminal Sca-1+ c-Kit+ cells, luminal genes Esr1 and Krt18 (b, c RHS), and basal gene Krt14 (d RHS) relative to comparator luminal Sca-1− c-Kit+/Low cells, in murine mammary epithelium. Data are presented as fold expression levels ±95% confidence intervals (n = three independently harvested isolates of each cell population). *Gene expression was undetectable in these populations in all three independent isolates. **Gene expression was only detected (at very low levels) in two of three isolates of the luminal Sca-1+ c-Kit− population. Therefore, no error bars are shown for this sample. Images used with permission under a CC-BY 4.0 license from Nguyen et al.51 and Regan et al.28.
Nguyen et al.51 observed fractions of cells that co-express both luminal K8 and basal K14 markers, and report that such K8+ K14+ cells had previously been observed in mouse fMASCs by Spike et al.37 (such fetal cells were also previously described by Sun et al.57), but not in adult human tissue in homeostasis. However, while the canonical view among mouse mammary developmental biologists is that the K5/14 pair is a basal marker and the K8/18 pair is a luminal marker58,59,60, breast pathologists have known for many years that keratins 5 and 14 (and indeed another “basal” keratin, 17) are in fact expressed in basal cells of human breast ducts and in the luminal cells of the terminal ductal lobuloalveolar units (TDLUs)58,61,62,63,64. Indeed, K5/K18 and K14/K18 double-positive cells are not uncommon in human TDLUs61. More recently, Boecker et al.65 identified K5+ K18/19− and K5+ K18/K19+ populations in the luminal layer of ductal and TDLU breast tissue in situ65, while in human breast epithelial populations isolated by flow cytometry, the progenitor populations (Lin− CD49f+ EpCAMhi) include cells double-positive for K5/6 and K14 — and notably are also c-KIT+40. To add to the complexity of these marker patterns, K19 has been described both as a marker of progenitors66,67,68 and highly expressed in differentiated luminal ER+ cells6,69.
Boecker et al.65 termed the populations they identified as progenitors and intermediary cells, respectively, but it is difficult to definitively assign such functions purely on the basis of marker expression, or indeed ex vivo assays. Of course, human breast tissue cannot be lineage-traced through transgene activation as one can in the mouse, but use of cytochrome C oxidase (CCO) mutations in the mitochondrial genome has proven feasible as an approach. Cereser et al.70 report the presence of CCO-deficient clonal expansions in both ducts and TDLUs of normal breast70. Notably, the expansions were limited to the luminal layers, and they found no evidence of luminal CCO-deficient clones contributing to the basal layer. Therefore, if the K5/K14/c-KIT+ luminal cells of the human breast are indeed progenitors, they are lineage-restricted.
Keratin expression patterns in the mouse mammary epithelium are somewhat easier to define, but also not as straightforward as often suggested. Unlike in the human, when analyzed in situ, K14 and K8/18 in the mouse appear to be restricted to the basal and luminal cell layers, respectively. Indeed, we have rarely (if ever) observed a luminal cell in the normal resting adult mammary gland we could confidently say is K14 positive, or a basal cell that is K8/18 positive, by immunofluorescence in situ, and this is in agreement with most studies. However, immunohistochemical analysis of the mouse mammary gland by Mikaelian et al.59 has detected rare weak K14 staining of luminal cells from birth to puberty and weak K8/18 labeling of basal cells during mammary morphogenesis, which were most easily visualized during lactation59. As an added complication, it should be noted that in the mammary alveoli, the basal/myoepithelial cells form a classic “basket-like network” around the secretory cells, and in that location, the “luminal” cells are in fact touching the basement membrane through the gaps between the myoepithelial cells. Interestingly, therefore, in agreement with Mikaelian et al.59, when basal and luminal subpopulations were isolated by flow cytometry and stained by immunofluorescence, we found that c-Kit+ luminal cells (which were approx. 50% of the total mammary epithelium) were all strongly K18+ but also weakly K14+, and that c-Kit+ basal cells were strongly K14+ and weakly K18+ (Fig. 2b)28. c-Kit-negative single luminal and basal cells prepared and stained at the same time were respectively K18+ K14− and K14+ K18−, suggesting that we were not seeing background staining in the c-Kit-positive cells. This discrepancy is likely due to signal/noise ratio when using in situ immunofluorescence approaches — enhancing the K14 staining to a level where it can be detected in luminal cells would result in a huge excess of staining from the basal cells as well as background signal from other cell types in the mammary gland (and likewise for K18 detection in basal cells), which is notorious for background fluorescence coming from adipocytes. Thus, only approaches based on single-cell separation will accurately detect mouse cells expressing the “luminal” keratin 18 and the “basal” keratin 14, and as we report using such approaches, such cells express the c-Kit marker28. Note that the scRNA-seq analysis of mouse mammary epithelium by Bach et al.53 shows that a subset of luminal cells have Krt14 expression levels equivalent to the mean expression level of Krt14 in basal cells. Their differentiation trajectory maps show that the Krt14-expressing luminal cells are enriched in a progenitor population that is also c-Kit-positive53.
a Immunofluorescence of sections though the mammary fat pads of adult virgin female FVB mice stained with antibodies against the luminal markers K18 and c-Kit and the basal marker K14. c-Kit staining is located predominantly in the K18+ K14− luminal layer, although occasional K14+ c-Kit+ basal cells are detected (arrowhead). Bar = 40 µm. b K18 and K14 staining of freshly isolated single c-Kit+ luminal and c-Kit+ basal cells from adult virgin mice sorted directly onto slides. Insets show c-Kit− luminal and basal cells negative for K14 (LHS) and K18 (RHS), respectively (bar = 3 µm). The numbers of cells examined and overall staining patterns are given in Table 1 of Regan et al.28. c Basal K5 staining in the terminal end buds (TEBs) and subtending duct of 4-week-old pubertal mouse mammary epithelium. K5 staining is located predominantly in the basal layer. Occasional K5+ cells are detected in the luminal layer (arrowheads). Bar = 40 μm. d Section through a cleared fat pad outgrowth double-stained for basal K5 and luminal K19. A K5+ K19+ double-positive cell is observed in the basal layer (arrowhead). Bar = 40 µm. All cells were counterstained with DAPI (blue).
In contrast, we find that cells double-positive for “basal” keratin 5 and “luminal” keratin 19 are readily detectable in the mouse luminal epithelium in situ (Fig. 2c, d). Interestingly, K19 has been proposed to be a neutral switch keratin that permits the changeover of one type of cytoskeleton to the other68,71. We have particularly noted K5-positive cells in the body cell region of terminal end buds in situ (Fig. 2c). The origin of these cells is unclear. Rios et al.16 reported that using a Krt5-promoter-driven cell-labeling approach, labeled cells were only observed in the basal compartment, but generated both luminal and basal daughter clones, and hence proposed the existence of bipotent basal stem cells arising from the basal layer of the TEBs16. However, the work of Scheele et al.23 and others18,19,20,21,22,23,46,47 suggests that cap cells (the basal cell layer of the TEBs) do not contribute to the luminal layer of the subtending duct; therefore K5-positive body cells, if they are cap cell-derived, are unlikely to contribute to outgrowth of the ducts. In contrast, if these cells are derived from the body cells, they are switching on high levels of K5 expression, but whether this is only transient — perhaps a temporary failure of lineage specification in a newly established daughter cell that is later corrected — is unclear.
Therefore, while use of keratins as basal/luminal lineage markers is more robust in the mouse mammary epithelium than in the human, single-cell analysis approaches have demonstrated that even the mouse has a more promiscuous pattern of keratin expression than previously suspected, and that this promiscuous expression of keratins is seen in c-KIT+ stem/progenitor cells. Plasticity in the expression of keratins and other genes within c-Kit+ luminal progenitors may relate to their potential to contribute to multiple cell lineages during epithelial remodeling, e.g., at involution of the mammary gland after weaning72. In addition, the phenotypic plasticity and multilineage differentiation potential of these luminal progenitors is consistent with their ability to give rise to tumors with basal features40,50, as well as lineage switching in response to injury and oncogene activation20,24,49. It is clear, therefore, that a great deal of caution must be used when keratin promoters are being used for lineage-tracing studies in the mouse or for assigning luminal/basal identity in human cells. Indeed, in a dissociated human breast epithelial cell population, keratin expression levels alone cannot be used to assign basal/luminal identity to a cell with any confidence.
To address the debate as to whether homeostasis and development in the postnatal mammary gland are maintained by bipotent MaSCs15,16,43 or lineage-restricted basal and luminal cells54,19,20,21,22, Nguyen et al.51 performed pseudotemporal reconstruction-based lineage hierarchy analysis. This analysis identified a continuous lineage connecting the basal lineage, via a bipotent MaSC, to the two luminal branches. These results agree with previous models of mammary differentiation wherein a bipotent basal MaSC generates daughter cells that differentiate into myoepithelial and luminal cell lineages15,16,43. However, Nguyen et al. propose that their results differ from previous studies in that L1.2 cells (luminal ER− c-kit+/Low cells) are progenitors to L1.1 cells (luminal ER− c-Kit+/High cells), and that c-Kit+/High L1.1 cells are another type of mature differentiated luminal cell rather than a luminal progenitor upstream of luminal ER+ L2 cells. Based on this pseudotemporal analysis, the authors suggest that KIT is not a marker of luminal progenitor cells. This is a surprising conclusion considering that L1.2 progenitor cells do express KIT (Fig. 1), which as well as being a defining marker of mouse and human progenitor cell gene expression signatures17,34,40,52,53,54,73, has been functionally demonstrated as a progenitor cell marker28 (Table 1).
Similar to Nguyen et al.51, Pal et al.52 used scRNA-seq to identify lineage relationships in the mouse mammary gland, and also suggested that bipotent basal MaSCs give rise to basal and luminal lineages52. Supporting our previous assessment of intermediate cells in the luminal lineage28, the authors also described the identification of intermediate luminal cells. Significantly, Pal et al. report the identification of rare mixed-lineage or “lineage-primed” c-Kit-expressing basal cells in the adult mammary gland and state, “It is presumed that these cells represent a transient population that is poised for commitment to the luminal lineage, reminiscent of “lineage-primed” stem and progenitor cells initially reported in the hematopoietic system.” These lineage-primed c-Kit+ basal cells comprised ~5% of the basal compartment and expressed luminal genes such as Esr1, Prlr, Csn2, and Areg in addition to basal genes. Pal et al. state, “these data suggest that the basal state may precede commitment to a luminal cell fate in the post-natal mammary gland.”
In Regan et al.28, we also identified cells that we described as lineage-primed basal cells (CD24+/Low Sca-1− CD49f+/High c-Kit+) in the adult mammary gland that expressed luminal genes, including those described by Pal et al. (Esr1, Prlr, Csn2, and Areg), but that clustered with the basal facultative MaSCs28. Significantly, we functionally tested these cells by single-cell cleared mammary fat pad transplantation and demonstrated that they can reconstitute an entire ductal tree, although at a lower frequency (1 in 8 ± 95% CI 1 in 3–1 in 21.3) than facultative c-Kit− MaSCs (1 in 3 ± 95% CI 1 in 1.69–1 in 6.27), the highest enrichment of facultative MaSCs reported to date and potentially a pure facultative MaSC population. Based on these data, we came to the same conclusion as Pal et al.52 and described these c-Kit+ basal cells as intermediate MaSCs that were undergoing “lineage priming,” in which stem cells express genes associated with their differentiated daughter populations74,75. This was the first time that lineage-primed basal cells in the adult mammary gland had been reported and functionally tested.
In contrast to Nguyen et al.51 and Pal et al.52, scRNA-Seq by Bach et al.53 on mouse mammary epithelial cells at nulliparous, mid gestation, lactation, and post involution concluded that, rather than clearly defined clusters maintained by their own stem/progenitor population, a continuous spectrum of differentiation exists. In this model, a common luminal progenitor cell, which notably expressed c-Kit at high levels, gives rise to intermediate, restricted alveolar, and hormone-sensitive progenitors.
More recently, Giraddi et al.54 used scRNA-seq and transposase-accessible chromatin sequencing (ATAC-seq), which examines global chromatin accessibility76 of embryonic, postnatal, and adult mouse mammary epithelia, to elucidate the lineage hierarchies and biological programs that generate mature cell types from their embryonic precursors54. This work was more consistent with the conclusions of Bach et al.53 than Nguyen et al.51 and Pal et al.52, as well as the lineage-tracing studies showing that while embryonic mammary cells are bipotent, in the adult gland, basal and luminal cell lineages are derived from and maintained by separate lineage-committed progenitor populations18,19,20,21,22,23,24,42,46,47,48.
Similar to Pal et al.52, Giraddi et al.54 also identified rare c-Kit+ basal cells, although they did not occur at a frequency greater than the expected doublet frequency (∼1%) of the 10X Genomics Chromium System sequencing platform54, a frequency similar to the c-Kit+ basal cells that Pal et al.52 also detected using the 10X platform. In contrast, the lineage-primed c-Kit+ basal cells that we identified in our 2012 study were visually confirmed to be single cells prior to performing the single-cell transplants, in which they displayed a transplantation-frequency intermediary to facultative c-Kit− MaSCs and c-Kit+ luminal progenitor cells. In addition, immunofluorescence staining of single c-Kit+ basal cells demonstrated that they expressed both K14 and K18 (Fig. 2b)28.
Transcriptional profiling by Giraddi et al.54 did not detect any distinct adult basal stem cell subpopulation. However, ATAC-seq revealed that adult basal cells display an embryonic MaSC-type chromatin accessibility at luminal gene loci, which the authors speculate allows for lineage plasticity54,73,77. Such plasticity may account for acquisition of multilineage potential upon perturbation of a homeostatic niche environment, such as during cell isolation and ex vivo culture, transplantation assays, wounding, and cancer49,54,77,78,79,80. The performance of a particular cell type during functional assays may therefore be a product of both their transcriptional heterogeneity and the context in which they are challenged49. Similar functional stem cell capacities have also been described in embryonic tissue, intestine, bone marrow, skin, and lung81,82,83. These observations challenge the concept of fixed-cell identities in complex tissues, and suggest a more fluid concept of cell state (for a more detailed discussion of this concept see Wahl and Spike49). With this in mind, a potential mammary epithelial cell hierarchy based on lineage tracing, functional analyse, and recent scRNA-seq and snATAC-seq studies is shown in Fig. 3.
Bipotent fetal mammary stem cells (fMaSCs) are present in the embryo and become lineage-restricted after birth. In the adult gland, each lineage is maintained by its own c-Kit+ progenitor. Loss of homeostasis (e.g., injury, cell isolation, ex vivo culture, and transplantation) or tumorigenesis may trigger a wound response that leads to acquisition of multilineage potential by facultative inducible MaSCs (iMaSCs), c-Kit+ lineage-primed, and progenitor cell states. Lineage-primed c-Kit+ basal cells that express intermediate levels of luminal genes may represent a transient or intermediate population that precedes commitment to the luminal lineage28,52. Gene expression analysis suggests that an alternative route for generating ER+ cells from intermediate luminal cell states may also exist.
Future studies that aim to map fluid cell-state dynamics and their regulatory mechanisms will require the use of single-cell and single-molecule epigenomic technologies that reveal a cell’s regulatory potential, rather than its current state, as indicated by its transcriptome84,85. Indeed, Chung et al.73 recently demonstrated that single-cell chromatin accessibility mapping of mammary gland development using single-nucleus ATAC-seq (snATAC-seq) enables greater resolution of cell-state heterogeneity, and to be a better indicator of cell state during development than scRNA-seq73. The lineage relationships delineated in this study were consistent with those of Bach et al.53 and Giraddi et al.54, and also found c-Kit to be most highly expressed and chromatin accessible in luminal progenitor cells.
Concluding remarks
Taken together, the weight of evidence supports c-Kit as a progenitor marker in the mammary epithelium and, more importantly, one that is functionally characterized and can be used to enrich stem/progenitor cells. Indeed, we have already begun to understand the signaling pathways downstream of c-Kit in mammary progenitor cells86. scRNA-seq studies, which allow for comprehensive and unbiased analysis of the different cell types that constitute a heterogeneous tissue87, have been extremely valuable in contributing to our understanding of lineage relationships and cell-state heterogeneity in the mammary gland. However, in order to fully understand the significance of these studies, it is essential to link them to functional data, in particular where such data already exist, and future studies should aim to do so. The evidence from lineage tracing, scRNA-seq, and snATAC-seq studies currently supports a model in which fMaSCs in the embryo are bipotent, whereas in the adult gland, stem/progenitor cells are lineage-restricted, and facultative MaSCs (defined by functional studies) are induced to acquire multilineage potential upon loss of homeostasis/injury. Bipotent fetal MaSCs are described as fMaSCs to differentiate them from adult facultative MaSCs. However, the scientific literature up to now continues to refer to adult cells with facultative stem cell potential simply as MaSCs or, in a handful of publications, adult MaSCs (aMaSCs)37,49, which is no longer an accurate or apt description. We therefore propose the renaming of MaSCs in the postnatal gland as “inducible mammary stem cells” (iMaSCs). This new definition will help to more clearly define the status and stem cell potential of functionally defined iMaSCs in the era of large-scale single-cell molecular profiling.
Data availability
Source data for all figures and tables are provided in the paper. No new datasets have been generated or analyzed for this article.
References
Regan, J. & Smalley, M. Prospective isolation and functional analysis of stem and differentiated cells from the mouse mammary gland. Stem Cell Rev. 3, 124–136 (2007).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006).
Plaks, V. et al. Lgr5 expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 3, 70–78 (2013).
Visser, K. E. et al. Developmental stage-specific contribution of LGR5+ cells to basal and luminal epithelial lineages in the postnatal mammary gland. J. Pathol. 228, 300–309 (2012).
Sleeman, K. E., Kendrick, H., Ashworth, A., I, C. M. & Smalley, M. J. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 8, R7 (2006).
Sleeman, K. E. et al. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J. Cell Biol. 176, 19–26 (2007).
Wilson, N. K. et al. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell 16, 712–724 (2015).
Smalley, M. J. et al. Isolation of mouse mammary epithelial subpopulations: a comparison of leading methods. J. Mammary Gland Biol. Neoplasia 17, 91–97 (2012).
Stingl, J. et al. Purification and unique properties of mammary epithelial stem cells. Nature 439, 993–997 (2006).
Shehata, M. et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res 14, R134 (2012).
Alvi, A. J. et al. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res. 5, R1–8 (2003).
Britt, K. L. et al. Pregnancy in the mature adult mouse does not alter the proportion of mammary epithelial stem/progenitor cells. Breast Cancer Res 11, R20 (2009).
Nguyen, L. V. et al. Clonal analysis via barcoding reveals diverse growth and differentiation of transplanted mouse and human mammary stem cells. Cell Stem Cell 14, 253–263 (2014).
Ginestier, C. et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of a poor clinical outcome. Cell Stem Cell 1, 555–567 (2007).
Wang, D. et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517, 81 (2014).
Rios, A. C., Fu, N. Y., Lindeman, G. J. & Visvader, J. E. In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322 (2014).
Lim, E. et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 12, R21–R21 (2010).
Chang, T. H.-T. et al. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res. 16, R1–R1 (2014).
Van Keymeulen, A. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189 (2011).
Van Keymeulen, A. et al. Lineage-restricted mammary stem cells sustain the development, homeostasis, and regeneration of the estrogen receptor positive lineage. Cell Rep. 20, 1525–1532 (2017).
Davis, F. M. et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat. Commun. 7, 13053 (2016).
Lloyd-Lewis, B., Davis, F. M., Harris, O. B., Hitchcock, J. R. & Watson, C. J. Neutral lineage tracing of proliferative embryonic and adult mammary stem/progenitor cells. Development 145, dev164079 (2018).
Scheele, C. L. G. J. et al. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature 542, 313–317 (2017).
Koren, S. et al. PIK3CAH1047R induces multipotency and multi-lineage mammary tumours. Nature 525, 114 (2015).
van Amerongen, R. in Mammary Stem Cells. Methods in Molecular Biology. (ed. Vivanco, M. del M.) 187–211 (Springer New York, 2015).
van de Moosdijk, A. A. A., Fu, N. Y., Rios, A. C., Visvader, J. E. & van Amerongen, R. in Mammary Gland Development. Methods in Molecular Biology, Vol. 1501. (eds. Martin, F., Stein, T. & Howlin, J.) 291–308 (Humana Press, New York, 2017).
DeOme, K. B., Faulkin, L. J. Jr., Bern, H. A. & Blair, P. B. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 19, 515–520 (1959).
Regan, J. L. et al. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene 31, 869 (2012).
Asselin-Labat, M. L. et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209 (2007).
Taddei, I. et al. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat. Cell Biol. 10, 716–722 (2008).
Jones, C. et al. Expression profiling of purified normal human luminal and myoepithelial breast cells: identification of novel prognostic markers for breast cancer. Cancer Res. 64, 3037–3045 (2004).
Grigoriadis, A. et al. Establishment of the epithelial-specific transcriptome of normal and malignant human breast cells based on MPSS and array expression data. Breast Cancer Res. 8, R56 (2006).
Shipitsin, M. et al. Molecular definition of breast tumor heterogeneity. Cancer Cell 11, 259–273 (2007).
Kendrick, H. et al. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genomics 9, 591 (2008).
Pece, S. et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 140, 62–73 (2010).
Wansbury, O. et al. Transcriptome analysis of embryonic mammary cells reveals insights into mammary lineage establishment. Breast Cancer Res. 13, R79–R79 (2011).
Spike, B. T. et al. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell 10, 183–197 (2012).
Raouf, A. et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 3, 109–118 (2008).
Pal, B. et al. Integration of microRNA signatures of distinct mammary epithelial cell types with their gene expression and epigenetic portraits. Breast Cancer Res. 17, 85 (2015).
Lim, E. et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913 (2009).
van Amerongen, R., Bowman, A. N. & Nusse, R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387–400 (2012).
Wuidart, A. et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cell Biol. 20, 666–676 (2018).
Regan, J. L. et al. Aurora A kinase regulates mammary epithelial cell fate by determining mitotic spindle orientation in a notch-dependent manner. Cell Rep. 4, 110–123 (2013).
Lopez-Garcia, C., Klein, A. M., Simons, B. D. & Winton, D. J. Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822 LP–825 (2010).
Ritsma, L. et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362–365 (2014).
Wang, C., Christin, J. R., Oktay, M. H. & Guo, W. Lineage-biased stem cells maintain estrogen-receptor-positive and -negative mouse mammary luminal lineages. Cell Rep. 18, 2825–2835 (2017).
Elias, S., Morgan, M. A., Bikoff, E. K. & Robertson, E. J. Long-lived unipotent Blimp1-positive luminal stem cells drive mammary gland organogenesis throughout adult life. Nat. Commun. 8, 1714 (2017).
Lilja, A. M. et al. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat. Cell Biol. 20, 677–687 (2018).
Wahl, G. M. & Spike, B. T. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. npj Breast Cancer 3, 14 (2017).
Molyneux, G. et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7, 403–417 (2010).
Nguyen, Q. H. et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun. 9, 2028 (2018).
Pal, B. et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun. 8, 1627 (2017).
Bach, K. et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun. 8, 2128 (2017).
Giraddi, R. R. et al. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 24, 1653–1666.e7 (2018).
Asselin-Labat, M.-L. et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol. Cell. Biol. 31, 4609–4622 (2011).
Stingl, J., Eaves, C. J., Zandieh, I. & Emerman, J. T. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res. Treat. 67, 93–109 (2001).
Sun, P., Yuan, Y., Li, A., Li, B. & Dai, X. Cytokeratin expression during mouse embryonic and early postnatal mammary gland development. Histochem. Cell Biol. 133, 213–221 (2010).
Dontu, G. & Ince, T. A. Of mice and women: a comparative tissue biology perspective of breast stem cells and differentiation. J. Mammary Gland Biol. Neoplasia 20, 51–62 (2015).
Mikaelian, I. et al. Expression of terminal differentiation proteins defines stages of mouse mammary gland development. Vet. Pathol. 43, 36–49 (2006).
Smith, G. H., Mehrel, T. & Roop, D. R. Differential keratin gene expression in developing, differentiating, preneoplastic, and neoplastic mouse mammary epithelium. Cell Growth Differ. 1, 161–170 (1990).
Santagata, S. et al. Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J. Clin. Invest. 124, 859–870 (2014).
Santagata, S. & Ince, T. A. Normal cell phenotypes of breast epithelial cells provide the foundation of a breast cancer taxonomy. Expert Rev. Anticancer Ther. 14, 1385–1389 (2014).
Gusterson, B. Do ‘basal-like’ breast cancers really exist? Nat. Rev. Cancer 9, 128–134 (2009).
Gusterson, B. A., Ross, D. T., Heath, V. J. & Stein, T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res. 7, 143–148 (2005).
Boecker, W. et al. Spatially correlated phenotyping reveals K5-positive luminal progenitor cells and p63-K5/14-positive stem cell-like cells in human breast epithelium. Lab. Investig. 98, 1065–1075 (2018).
Clarke, R. B. et al. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev. Biol. 277, 443–56 (2005).
Gudjonsson, T. et al. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 16, 693–706 (2002).
Villadsen, R. et al. Evidence for a stem cell hierarchy in the adult human breast. J. Cell Biol. 177, 87–101 (2007).
Bartek, J., Bartkova, J. & Taylor-Papadimitriou, J. Keratin 19 expression in the adult and developing human mammary gland. Histochem. J. 22, 537–544 (1990).
Cereser, B. et al. Analysis of clonal expansions through the normal and premalignant human breast epithelium reveals the presence of luminal stem cells. J. Pathol. 244, 61–70 (2018).
Stasiak, P. C., Purkis, P. E., Leigh, I. M. & Lane, E. B. Keratin 19: predicted amino acid sequence and broad tissue distribution suggest it evolved from keratinocyte keratins. J. Invest. Dermatol. 92, 707–716 (1989).
Wagner, K. U. et al. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development 129, 1377–1386 (2002).
Chung, C. Y. et al. Single-cell chromatin analysis of mammary gland development reveals cell-state transcriptional regulators and lineage relationships. Cell Rep. 29, 495–510.e6 (2019).
Huang, S., Guo, Y. P., May, G. & Enver, T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 305, 695–713 (2007).
Månsson, R. et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity 26, 407–419 (2007).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213 (2013).
Dravis, C. et al. Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34, 466–482.e6 (2018).
Ge, Y. et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650.e14 (2017).
Ge, Y. & Fuchs, E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat. Rev. Genet. 19, 311 (2018).
Seldin, L., Le Guelte, A. & Macara, I. G. Epithelial plasticity in the mammary gland. Curr. Opin. Cell Biol. 49, 59–63 (2017).
Blanpain, C. & Fuchs, E. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281–1242281 (2014).
Hough, S. R., Laslett, A. L., Grimmond, S. B., Kolle, G. & Pera, M. F. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS ONE 4, e7708 (2009).
Tata, P. R. & Rajagopal, J. Plasticity in the lung: making and breaking cell identity. Development 144, 755–766 (2017).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193 (2016).
Shema, E., Bernstein, B. E. & Buenrostro, J. D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 51, 19–25 (2019).
Tornillo, G. et al. Dual mechanisms of LYN kinase dysregulation drive aggressive behavior in breast cancer cells. Cell Rep. 25, 3674–3692.e10 (2018).
Cristea, S. & Polyak, K. Dissecting the mammary gland one cell at a time. Nat. Commun. 9, 2473 (2018).
Natali, P. G. et al. Expression of c-kit receptor in normal and transformed human nonlymphoid tissues. Cancer Res. 52, 6139 LP–6143 (1992).
Matsuda, R. et al. Expression of the c-kit protein in human solid tumors and in corresponding fetal and adult normal tissues. Am. J. Pathol. 142, 339–346 (1993).
Hines, S. J., Organ, C., Kornstein, M. J. & Krystal, G. W. Coexpression of the c-kit and stem cell factor genes in breast carcinomas. Cell Growth Differ. 6, 769–779 (1995).
Ulivi, P. et al. c-kit and SCF expression in normal and tumor breast tissue. Breast Cancer Res. Treat. 83, 33–42 (2004).
Tsuda, H. et al. Frequent KIT and epidermal growth factor receptor overexpressions in undifferentiated-type breast carcinomas with ‘stem-cell-like’ features. Cancer Sci. 96, 333–339 (2005).
Kim, J. & Villadsen, R. Expression of luminal progenitor marker CD117 in the human breast gland. J. Histochem. Cytochem. 66, 879–888 (2018).
Westbury, C. B. et al. Genome-wide transcriptomic profiling of microdissected human breast tissue reveals differential expression of KIT (c-Kit, CD117) and oestrogen receptor-α (ERα) in response to therapeutic radiation. J. Pathol. 219, 131–140 (2009).
Acknowledgements
The authors thank Geoffrey M. Wahl (The Salk Institute for Biological Studies) for his critical reading of the paper and helpful comments.
Author information
Authors and Affiliations
Contributions
Conceptualization and writing of the original draft, J.L.R.; review and editing, J.L.R. and M.J.S. All the authors read and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Regan, J.L., Smalley, M.J. Integrating single-cell RNA-sequencing and functional assays to decipher mammary cell states and lineage hierarchies. npj Breast Cancer 6, 32 (2020). https://doi.org/10.1038/s41523-020-00175-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-020-00175-8
This article is cited by
-
Defining mammary basal cell transcriptional states using single-cell RNA-sequencing
Scientific Reports (2022)
-
Single-cell evaluation reveals shifts in the tumor-immune niches that shape and maintain aggressive lesions in the breast
Nature Communications (2021)
-
Characterization of Gene Expression Signatures for the Identification of Cellular Heterogeneity in the Developing Mammary Gland
Journal of Mammary Gland Biology and Neoplasia (2021)