Abstract
Coronary artery disease is an inflammatory disorder characterized by narrowing of coronary arteries due to atherosclerotic plaque formation. To date, the accumulated epidemiological evidence supports an association between oral bacterial diseases and coronary artery disease, but has failed to prove a causal link between the two. Due to the recent surge in microbial identification and analyses techniques, a number of bacteria have been independently found in atherosclerotic plaque samples from coronary artery disease patients. In this study, we present meta-analysis from published studies that have independently investigated the presence of bacteria within atherosclerotic plaque samples in coronary artery disease patients. Data were collated from 63 studies covering 1791 patients spread over a decade. Our analysis confirms the presence of 23 oral commensal bacteria, either individually or in co-existence, within atherosclerotic plaques in patients undergoing carotid endarterectomy, catheter-based atherectomy, or similar procedures. Of these 23 bacteria, 5 (Campylobacter rectus, Porphyromonas gingivalis, Porphyromonas endodontalis, Prevotella intermedia, Prevotella nigrescens) are unique to coronary plaques, while the other 18 are additionally present in non-cardiac organs, and associate with over 30 non-cardiac disorders. We have cataloged the wide spectrum of proteins secreted by above atherosclerotic plaque-associated bacteria, and discuss their possible roles during microbial migration via the bloodstream. We also highlight the prevalence of specific poly-microbial communities within atherosclerotic plaques. This work provides a resource whose immediate implication is the necessity to systematically catalog landscapes of atherosclerotic plaque-associated oral commensal bacteria in human patient populations.
Similar content being viewed by others
Background
Human microbiome is now recognized as a central player in human health, and the oral microbiome is of increasing significance in human biology. The oral cavity includes several microbial habitats and is the central channel for entry of bacteria into the human body. It constitutes the second most complex ecological system in human body after the gut microbiome, with over 700 species of bacteria and >5 billion bacteria.1 These gram-positive or gram-negative bacteria colonize periodontal surfaces and are part of the saliva. Further, depending on oxygenation conditions, they may be aerobic (obligates or facultative) or anaerobic (obligates or facultative). In healthy conditions, the oral bacterial community of ~700 species is stable but remains susceptible to alteration in its population structures due to infections or other stresses (Human Oral Microbiome Database). Increasing evidence on known molecular diversity of bacteria and recent advances in culture-independent techniques has validated the involvement of oral microbiome in several autoimmune and metabolic events such as obesity, diabetes and cardiovascular diseases.2,3,4,5
In 2012, an estimated 17.5 million people died from cardiovascular diseases, of which 7.4 million deaths were due to coronary artery disease (CAD) arising from atherosclerosis.6 CAD is a chronic inflammatory disorder characterized by narrowing of the coronary artery due to plaque formation, and results in blocking or reducing oxygen-rich blood supply to the heart that may subsequently cause myocardial infarction.7 The major content of an atherosclerotic plaque is atheroma that is composed of macrophages, cholesterol, smooth muscles and dystrophic calcification.8 Interestingly, with the development of targeted microbial techniques a number of oral bacteria have been identified in atherosclerotic plaque samples.
The link between dental disease and CAD was first established ~23 years ago when De Stefano et al. reported an increased risk of atherosclerotic plaque formation in a group of patients with periodontitis (25 % higher) based on 14 years of research on 9760 individuals aged between 25–74 years.9 More recent studies have correlated oral microbial dysbiosis/infections with obesity as well as diabetes, two known drivers of CAD.5 The oral route is a key avenue for entry of bacteria into the human body, and the prevailing hypothesis for the above-established link suggests that there is a flow of bacterial toxins and/or bacterial components into the bloodstream, leading to exaggerated release of inflammatory mediators that can drive CAD.10 For example, it has been demonstrated that lipopolysaccharide (LPS)-derived products released during endotoxemia are contributors in the host–bacteriadialog, whereas LPS increases serum cholesterol levels by increasing low-density lipoprotein (LDL) thereby increasing the risk of developing CAD.11 Similarly, other bacteria-derived components such as DNA or membrane phospholipids from oral cavity may also reach the blood stream, and finally into adipose and vascular tissues. Thus, the oral cavity may represent a significant source of bacterial mediators (direct or indirect) that may have an impact on CAD.
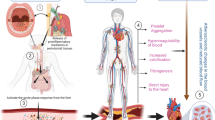
In order to assess the role of oral bacteria as inflammatory stimuli, it is important to understand the contribution of human immune system toward the formation of atherosclerotic plaque. The process of plaque formation in coronary arteries is initiated by the accumulation of LDL across the endothelium, leading to its retention in intima.12, 13 Following this deposition, LDL undergoes oxidative modification via enzymes secreted by endothelial cells.14 Oxidized LDL (OxLDL) trapped in the intima stimulates endothelial cells to secrete pro-inflammatory molecules, chemotactic proteins and growth factors that help in the recruitment of monocytes to the arterial wall and promote their differentiation and proliferation to macrophages.15 Macrophages then adhere to the endothelium and phagocytose OxLDL, leading to the formation of foam cells. The continued damage to endothelial cells leads to secretion of growth-stimulating factors, which in turn stimulates the proliferation of macrophages to function as foam cells.16 These foam cells then rupture and accumulate on the arterial wall. Following the deposition of lipids within the arterial wall, immune cells such as monocytes, leukocytes, B-cells, T-cells, neutrophils, dendritic cells (DC) and mast cells are recruited, which is the hallmark of atherosclerotic plaque formation17,18,19,20,21,22 (Fig. 1).
Schematic representation of inflammatory mechanisms involved in pathogenesis of atherosclerosis and plaque formation. LDL is retained in arterial intima via ionic interactions with endothelial cells, leading to the enzymatic oxidative modification of LDL into OxLDL. This is followed by secretion of pro-inflammatory cytokines that leads to the differentiation of monocytes into macrophages. Macrophages secrete more chemokines and mediate recruitment of neutrophils via scavenger receptors and further attract monocytes. Macrophages retained in arterial intima get converted into foam cells leading to the formation of atherosclerotic plaques
Atherosclerotic plaque formation may also be initiated by oral microbial dysbiosis/infection resulting in an inflammatory stimulus. For example (1) chronic periodontitis, a well-studied oral microbial disease with immunological implications, begins as an inflammation localized to the soft tissues (gingivitis) caused by resident biofilm that forms on tooth surfaces at the gingival margin. If left untreated, this leads to damage of connective tissue, periodontal ligament and bone.23, 24 (2) Gingival ulceration in periodontitis results in bacteraemia and can provide an additional inflammatory stimulus for atherosclerotic plaque formation.25, 26 The inflammatory cytokines produced and additional chemotactic agents lead to changes in the endothelium, e.g. via upregulation of adhesion molecules. These changes promote interactions with leukocytes, further promoting their migration into the intimal layer of the artery. Activation of the endothelium also results in the release of chemotactic cytokines, further attracting monocytes or other cells that form a vicious cycle leading to plaque formation.8
The aims of our study were to (i) collate and analyze associations between atherosclerotic plaque-associated bacteria in CAD patients from numerous independent studies, (ii) determine non-cardiac distribution of the oral bacteria in human body, (iii) dissect probable entry routes of oral bacteria into the coronary vasculature, (iv) highlight the plethora of proteins and peptides that are secreted by these bacteria, and finally (v) analyze the establishment of poly-bacterial communities within the plaques (Table 1).
Results
Distribution of oral bacteria in atherosclerotic plaques
Our literature search resulted in the selection of 63 studies and the identification of 23 bacteria that individually or otherwise co-existed in the studied human atherosclerotic plaque samples. Techniques used for the identification of bacteria within the atherosclerotic plaque samples included traditional methods like (1) categorizing on the basis of morphological characteristics, (2) culturing and isolation of colonies from samples. In addition, more modern techniques like immunohistochemistry, immunofluorescence, real-time polymerase chain reaction (PCR), nested PCR, and 16S rRNA gene sequencing were also used to identify bacteria. Among these techniques, 16S rRNA gene sequencing is the most reliable, cost effective, and scalable method when studying a large group of samples (Fig. 2). Our analyses show that 16S rRNA gene sequencing was the dominant technique (used 48 times) over traditional PCR methods (29 times) and other techniques like immunofluorescence and immune-histochemistry (15 times). Specifically, only 16S rRNA gene sequencing methodology identified Streptococcus sp., Enterobacter hormaechei, Pseudomonas aeruginosa, Pseudomonas luteola, Veillonella (Fig. 2a), and overall it identified 16 of the 23 atherosclerotic plaque-associated bacteria in CAD patients (Figs. 2b–d).
a Graphical representation of the techniques used to identify the atherosclerotic plaque-associated bacteria in CAD patients. Y-axis represents the number of studies reporting the presence of bacteria in the plaque samples, while X-axis depicts the corresponding bacteria. The cohort of 23 commensal bacteria is dominated by gram-negative bacteria with the exception of Streptococcus sp., which are gram-positive. Of these 23 atherosclerotic plaque-associated bacteria, A. actinomycetemcomitans, C. rectus, E. corrodens, E. hormaechei, S. gordonii, S. mitis, S. mutans, S. oralis, S. sanguinis, H. pylori, and P. aeruginosa are facultative anaerobes, while C. pneumoniae, F. necrophorum, F. nucleatum, M. pneumoniae, P. endodontalis, P. gingivalis, P. intermedia, P. nigrescens, T. denticola, and T. forsythia are obligatory anaerobes. There are two exceptions in P. luteola (aerobe) and Veillonella (anaerobe). IF immunofluorescence, IHC immunohistochemistry, mAb monoclonal antibodies. b Atherosclerotic plaque-associated bacteria identified using 16S rRNA technique. X-axis represents the % of patients positive for the bacteria identified using 16S rRNA gene sequencing, while Y-axis represents the corresponding bacteria. Total number of study subjects vs. positive patients is mentioned on the top of graph. c Atherosclerotic plaque-associated bacteria identified using traditional PCR techniques. X-axis represents the % of patients positive for the bacteria identified using traditional PCR techniques, while Y-axis represents the corresponding bacteria. Total number of study subjects vs. positive patients is mentioned on the top of graph. d Atherosclerotic plaque-associated bacteria identified using immunofluorescence, immuno-histochemistry, and antibody screening methods. X-axis represents the % of patients positive for the bacteria identified using multiple techniques, while Y-axis represents the corresponding bacteria. Total number of study subjects vs. positive patients is mentioned on the top of graph
Atherosclerotic plaque-associated bacteria in non-cardiac organs
On investigating the tissue distribution of 23 atherosclerotic plaque-associated bacteria, we found that some of these were found in several body organs (Fig. 3). The translocation of these bacteria into the bloodstream and subsequently to multiple organs may be triggered by tissue damage via periodontal probing, scaling, and tooth extractions, and/or aided by the proteins secreted by these bacteria. For example, Chlamydiae pneumoniae can lead to chronic obstructive pulmonary disease,27 sexually acquired reactive arthritis,28 asthma,29 increase the risk of developing lung cancer, and is present within brain regions of Alzheimer patients.30 In addition, C. pneumoniae reinfection accelerates the development of insulin resistance and diabetes in obese C57BL/6 mice.31 Similarly, Aggregatibacter actinomycetemcomitans is responsible for brain abscess, infectious arthritis,32 rib destruction33 as well as infective endocarditis.34 Of the 23 bacteria, Fusobacterium nucleatum is associated with inflammatory bowel disease,35 ulcerative colitis,36 and intestinal tumorigenesis.37, 38 The most studied of these are P. gingivalis and Helicobacter pylori and their detailed associations are depicted and categorized on the basis of tissue or organ system affected (Fig. 4). These data together highlight the complex associative underpinnings of oral commensals and human body organs.
The tissue localization of the 23 oral commensal bacteria associated with atherosclerotic plaque samples from CAD patients. Sixteen of the 23 atherosclerotic plaque-associated bacteria were not unique to atherosclerotic plaque samples and are present in multiple non-cardiac organs (gram-negative microbes are in red)
Proteins/peptides predicted to be secreted by atherosclerotic plaque-associated bacteria
We cataloged 36 predicted secretory proteins from 16 plaque-associated bacteria (Table 2). These proteins likely have multiple functions that include aiding in bacterial pathogenesis, increasing the virulence of the bacteria, and/or regulating host immune responses. Further, we analyzed these 36 secretory proteins in context of the host immune system and categorized them on the basis of their potential to influence oral cavity and immune system that could lead to inflammation (Fig. 5a). Both H. pylori and P. gingivalis have been studied extensively in context of their involvement in various disease pathologies. Secretory proteins such as gingipains from P. gingivalis and Hpn from H. pylori are known to activate cytokine secretion (mainly IL-6 and IL-8).39,40,41 Similarly, the CD-14 binding protein of S. sanguinis also results in secretion of host cytokines IL-6 and IL-8.42 Further, the protein SerbB secreted by P. gingivalis modulates host cytoskeleton, thus aiding microbes to enter host tissues.43 The leukotoxin (Lkt A) secreted by Fusobacterium necrophorum,44 Dup A,45 HtrA46 of H. pylori, and interpain A of P. intermedia 47 all serve as virulence factors for their respective bacteria, assisting them in infecting the host cells. The elastase A (Las A) and elastase B (Las B) of P. aeruginosa also have probable roles in bacterial pathogenesis.48
Proteins secreted by the atherosclerotic plaque-associated bacteria. a Histogram representing the number of secretory protein/peptides and proteases from atherosclerotic plaque-associated bacteria. b The gingival crevice is a habitat to many oral microbes that secrete proteins, peptides and proteases. (1) Secretory peptides and proteases are likely responsible for altering the host actin cytoskeleton in the gingival epithelium leading to microbial entry into the system. (2) These secreted proteins can also activate the immune system causing inflammation. Primarily, cytokine-mediated (IL-6 and IL-8) inflammation is associated with atherosclerotic plaque formation. Certain proteases cause inflammatory response by activating the complement system
We present a model for possible access routes of bacteria into the epithelial tissues (Fig. 5b). When in blood, commensal bacteria can invade the endothelial layer of the blood vessels with help of secretory proteins, and stimulate the production of pro-inflammatory cytokines such as monocyte chemo-attractant protein 1, IL-6, and IL-8.49, 50 These inflammatory cytokines can result in recruitment of DC, which then phagocytose oral bacteria and carry them through the blood stream until they are deposited in the vascular sites.51 This thus provides a potential entry point for the oral bacteria, and enables their migration from oral cavity into the blood stream—and feasibly to the coronary arterial system. In addition to the above, several proteins secreted by oral bacteria (Table 2) are capable of degrading oral mucosal membranes and periodontal pockets, again facilitating the entry of bacteria into the blood stream. It remains unclear whether oral bacteria nucleate atherosclerotic plaque formation or that they are deposited in the plaque site once it has developed.
Poly-microbial community in atherosclerotic plaques
The survival strategies adopted by commensal bacteria are of interest as they allow formation of a poly-microbial environment.52 With the exception of aerobe P. luteola, the other 22 atherosclerotic plaque-associated bacteria are either facultative or obligatory anaerobes. Under aerobic conditions, anaerobic bacteria have been shown to form biofilm structures, thus establishing an intricate and genetically varied microenvironment to survive.53 The process of biofilm formation in the oral cavity is initiated with the aggregation of early colonizers like Actinomycetes, Streptococcus and Veillonella. The Actinomycetes and Streptococcus sp. are present in almost equal ratio during the initial stages of biofilm formation in oral cavity.54,55,56 The Streptococcus and Actinomyces interact such that cell wall polysaccharide of Streptococcus binds with Type II fimbriae of Actinomyces resulting in first step towards biofilm formation.57 Further, the metabolic products of Streptococcus sp. such as lactic and pyruvic acid are exploited by Actinomycetes and Veillonella to support their own growth. Contrary to this, Streptococcus sp. convert excess lactic acid to hydrogen peroxide, thus preventing the attachment and growth of other periodontal pathogens.58 In the next phase of biofilm formation, F. nucleatum acts as a middle colonizer—a bridge between early and late colonizers59—and facilitates the adhesion of early colonizer Streptococcus sp. via an adhesion protein called RadD.60 Simultaneously, F. nucleatum provides its serotype and lecitin-carbohydrate-specific adhesins to the late colonizers—P. ginigvalis, A. actinomycetemcomitans and Treponema denticola.61, 62 Similar to early colonizers, the late colonizers also show co-adherence among themselves. For example, T. denticola secretes chymotrypsin-like proteinases that aid in adhering to the existing late colonizers in order to form polymicrobial community.63 Further, succinate formed by T. denticola is exploited by P. ginigvalis, which in turn promotes the growth of T. denticola by providing isobutyric acid52 (Fig. 6).
Atherosclerotic plaque-associated bacteria form biofilm structures within the atherosclerotic plaque samples. During initial phase of biofilm formation, early colonizers—Veillonella, Streptococcus, and Actinomyces—interact to establish an initial microenvironment supporting each other with the help of metabolic products. These bacteria act as a platform for the middle colonizer F. nucleatum, which then completes the biofilm formation by providing an adhering platform for the late colonizers—T. forsythia, A. actinomycetemcomitans, T. denticola, and P. gingivalis
In addition to the above-listed resident colonizers, both commensal and pathogenic bacteria have been shown to form biofilm structures. For instance, P. intermedia is a commensal to the healthy gingival crevices,64 while P. gingivalis is responsible for its invasion, resulting in the periodontal disease.65 Once within the gingival crevices, P. gingivalis aids P. intermedia to form biofilm structures with the help of virulence factors like arginine (Rgp) and lysine-specific cysteine proteases (Kgp).66 Simultaneously, Porphyromonas ginigvalis acts to detach A. actinomycetemcomitans from within the biofilms with the help of Kgp.67 Thus, bacteria can invade healthy gingival crevices by detaching and distorting the already existing oral biofilm. This can damage connective tissue, periodontal ligament, and bone with the help of bacterially secreted peptides and proteases, thereby allowing the bug access to bloodstream. Upon gaining entry into the coronary vasculature, these migratory bacteria can form biofilm structures within atherosclerotic plaques. For example, F. nucleatum and Streptococcus sp. forms a corncorb-like structure within the human atherosclerotic plaque.68, 69 Thus, these atherosclerotic plaque-associated bacteria may form mutually beneficial poly-microbial communities.70
Discussion
The oral cavity is a complex part of the human system and a number of factors work in synergy to maintain its homoeostasis. The oral system serves as a major route for the entry of bacteria to populate and establish a microenvironment within the human system. In this respect, the oral commensal bacterial species not only maintain harmony within themselves via formation of biofilms and polymicrobial communities, but also with the host body by educating the immune system and contributing toward health.71, 72 The compositions of saliva and commensal bacterial populations in the oral cavity are inter-related, and over 700 bacterial species are housed.71, 72 For instance, the host glycoprotein component of saliva provides nutrition to the oral bacteria, whereas antimicrobials peptides secreted by the host system present in saliva keep the oral microbial populations in-check.73 Alternatively, the oral bacteria secrete proteins that degrade host defense peptides (anti-microbial) in saliva to sustain in the oral cavity.74, 75 Hence, the oral microbiome is increasingly considered a very significant player in human health and disease. In this study, we have shown the potential of the 23 atherosclerotic plaque-associated oral commensal bacteria in disease pathology. The cohort of 23 atherosclerotic plaque-associated bacteria is dominated by gram-negative bacteria with the exception of Streptococcus sp. Full genomes of 19 of the 23 bacteria from this cohort are now available, except E. corrodens, E. hormaechei, F. necrophorum, and P. nigrescens. Hence, a genomic platform has been established to enable bacterial and molecular profiling of factors that contribute to plaque formation. Further investigations of these microbe–plaque axes are now required to unravel the full extent of linkage between host microbiome with atherosclerosis.
Methods
Data collection
Data sets selected in this study were sourced from published material from PUBMED, ATCC, and online web sources (Kenyon Microbe Wiki and Google search). Using a cataloguing procedure described in Fig. 7, we annotated all known bacteria that have been identified from atherosclerotic plaque samples of CAD patients. In brief, all PUBMED titles and abstracts were screened for eligibility. Any pre-clinical study was eligible for inclusion if it reported data regarding the presence of certain bacteria in atherosclerotic plaque within a coronary artery. These studies included randomized controlled trials, prospective case series, and controlled studies. Single case reports, conference proceedings, abstracts, and letters to the editor were screened but excluded if essential methodological information was missing. Additional articles from cross-references, which were missed due to absence of the search terms in title or abstracts, were hence included. Our literature search therefore resulted in selection of 63 studies, and identification of 23 bacteria in the human atherosclerotic plaque samples.
Study selection criteria to determine the population of microbes present in atherosclerotic plaques of CAD patients. Cataloguing procedure used to annotate all known oral microbes that have been identified from atherosclerotic plaque samples of CAD patients. In brief, the records present in PUBMED were studied by using combination of terminologies such as microbes, microorganism, and bacteria along with CAD, cardiovascular disease, atheroma, atherosclerosis, and atherosclerotic plaque. The articles were collated, duplicates removed, and relevant data extracted
Characterization on the basis of tissue localization and disease caused
Data sets selected in this study were sourced from published material from PUBMED, ATCC, and online web sources (Kenyon Microbe Wiki and Google search). In brief, all PUBMED titles and abstracts were screened for tissue localizations and diseases caused by the 23 atherosclerotic plaque-associated bacteria. Using above data, a map showing the presence of bacteria in various organ systems was generated. A GG plot of bacteria and diseases caused by them was prepared in R programming language.
Identification of secretory proteins
The data pertinent in cataloguing secretory proteins/peptides from the 23 atherosclerotic plaque-associated bacteria were sourced from PUBMED. The screening process included only those studies that incorporated proper characterization of proteins/peptides secreted by any of the microbes in focus here. This resulted in a total of 33 articles whose data were added to this study.
References
Paster, B., Olsen, I., Aas, J. & Dewhirst, F. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 42, 80–87 (2006).
Turnbaugh, P. J., Bäckhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Abusleme, L. et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025 (2013).
Serra e Silva Filho, W. et al. Microbial diversity similarities in periodontal pockets and atheromatous plaques of cardiovascular disease patients. PLoS ONE 9, e109761 (2014).
Shillitoe, E. et al. The oral microflora in obesity and type-2 diabetes.J. Oral Microbiol. 4, doi:10.3402/jom.v4i0.19013 (2012).
World Health Organization. Cardiovascular diseases (CVDs). Fact sheet No.317, January 2015 4–7 (2015).
Weber, C. & Noels, H. Atherosclerosis: current pathogenesis and therapeutic options. Nat. Med. 17, 1410–1422 (2011).
Bentzon, J. F., Otsuka, F., Virmani, R. & Falk, E. Mechanisms of plaque formation and rupture. Circ. Res. 114, 1852–1866 (2014).
DeStefano, F., Anda, R. F., Kahn, H. S., Williamson, D. F. & Russell, C. M. Dental disease and risk of coronary heart disease and mortality. Br. Med. J. 306, 688–691 (1993).
Han, Y. W. & Wang, X. Mobile microbiome: Oral bacteria in extra-oral infections and inflammation. J. Dent. Res. 92, 485–491 (2013).
Feingold, K. R. et al. Effect of endotoxin on cholesterol biosynthesis and distribution in serum lipoproteins in Syrian hamsters. J. Lipid. Res. 34, 2147–2158 (1993).
Nievelstein, P. F., Fogelman, A. M., Mottino, G. & Frank, J. S. Lipid accumulation in rabbit aortic intima 2 hours after bolus infusion of low density lipoprotein. A deep-etch and immunolocalization study of ultrarapidly frozen tissue. Arterioscler. Thromb. Vasc. Biol. 11, 1795–1805 (1991).
Tabas, I., Williams, K. J. & Borén, J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: Update and therapeutic implications. Circulation 116, 1832–1844 (2007).
Perrin-Cocon, L. et al. Oxidized low-density lipoprotein promotes mature dendritic cell transition from differentiating monocyte. J. Immunol. 167, 3785–3791 (2001).
Catapano, A. L., Maggi, F. M. & Tragni, E. Low density lipoprotein oxidation, antioxidants, and atherosclerosis. Curr. Opin. Cardiol. 15, 355–363 (2000).
Kusumoto, F. M. Cardiovasc. Pathophysiol. Hayes Barton Press, Raleigh, North Carolina (2004).
Lusis, A. Atherosclerosis. Nature 407, 233–241 (2000).
Gerrity, R. G., Naito, H. K., Richardson, M. & Schwartz, C. J. Dietary induced atherogenesis in swine. Morphology of the intima in prelesion stages. Am. J. Pathol. 95, 775–792 (1979).
Weber, C., Zernecke, A. & Libby, P. The multifaceted contributions of leukocyte subsets to atherosclerosis: Lessons from mouse models. Nat. Rev. Immunol.8, 802–815 (2008).
Ley, K. & Galkina, E. Leukocyte influx in atherosclerosis. Curr. Drug Targets 8, 1239–1248 (2007).
Hansson, G. K. & Libby, P. The immune response in atherosclerosis: A double-edged sword. Nat. Rev. Immunol. 6, 508–519 (2006).
Soehnlein, O. Multiple roles for neutrophils in atherosclerosis. Circ. Res. 110, 875–888 (2012).
Igari, K., Kudo, T., Toyofuku, T., Inoue, Y. & Iwai, T. Association between periodontitis and the development of systemic diseases. Oral Biol. Med. 2, 4 (2014).
Khalaf, H., Lönn, J. & Bengtsson, T. Cytokines and chemokines are differentially expressed in patients with periodontitis: Possible role for TGF-β1 as a marker for disease progression. Cytokine 67, 29–35 (2014).
Bartova, J. et al. Periodontitis as a risk factor of atherosclerosis. J. Immunol. Res. doi:10.1155/2014/636893 (2014).
Iwai, T. Periodontal bacteremia and various vascular diseases. J. Periodontal. Res. 44, 689–694 (2009).
Beaty, C. D. et al. Chlamydia pneumoniae, strain TWAR, infection in patients with chronic obstructive pulmonary disease. Am. Rev. Respir. Dis. 144, 1408–1410 (1991).
Bas, S. et al. Synovial fluid and serum antibodies against chlamydia in different forms of arthritis: Intra-articular IgA production in chlamydia sexually acquired reactive arthritis. Br. J. Rheumatol. 35, 548–552 (1996).
Hahn, D. L. et al. Chlamydia pneumoniae-specific IgE is prevalent in asthma and is associated with disease severity. PLoS ONE, 7, e35945 (2012).
Balin, B. J. et al. Identification and localization of chlamydia pneumoniae in the alzheimer’s brain. Med. Microbiol. Immunol. 187, 23–42 (1998).
Wang, C., Gao, D. & Kaltenboeck, B. Acute chlamydia pneumoniae reinfection accelerates the development of insulin resistance and diabetes in obese C57BL/6 mice. J. Infect. Dis. 200, 279–287 (2009).
Molina, F., Echániz, A., Durán, M. T. & Diz-Lois, F. Infectious arthritis of the knee due to actinobacillus actinomycetemcomitans. Eur. J. Clin. Microbiol. Infect. Dis. 13, 687–689 (1994).
Yuan, A. et al. Actinobacillus actinomycetemcomitans pneumonia with chest wall involvement and rib destruction. Chest 101, 1450–1452 (1992).
Das, M., Badley, A. D., Cockerill, F. R., Steckelberg, J. M. & Wilson, W. R. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 48, 25–33 (1997).
Strauss, J. et al. Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel. Dis. 17, 1971–1978 (2011).
Tahara, T. et al. Fusobacterium detected in colonic biopsy and clinicopathological features of ulcerative colitis in Japan. Dig. Dis. Sci. 60, 205–210 (2014).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Flanagan, L. et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1381–1390 (2014).
Jayaprakash, K., Khalaf, H. & Bengtsson, T. Gingipains from porphyromonas gingivalis play a significant role in induction and regulation of CXCL8 in THP-1 cells. BMC Microbiol. 14, 193 (2014).
Lourbakos, A. et al. Arginine-specific protease from porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion arginine-specific protease from porphyromonas gingivalis activates protease-activated recept. Infect. Immun. 69, 5121–5130 (2001).
Zhou, Q., Qi, S., Sun, X. & Ge, R. The interaction of a histidine-rich protein Hpn with the membrane mimics: Implications for pathologic roles of Hpn in helicobacter pylori. Helicobacter 19, 129–135 (2014).
Banks, J. et al. Streptococcus sanguis secretes CD14-binding proteins that stimulate cytokine synthesis: A clue to the pathogenesis of infective (bacterial) endocarditis?. Microb. Pathog. 32, 105–116 (2002).
Hasegawa, Y. et al. Role of porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect. Immun. 76, 2420–2427 (2008).
Narayanan, S. K., Nagaraja, T. G., Chengappa, M. M. & Stewart, G. C. Cloning, sequencing, and expression of the leukotoxin gene from fusobacterium necrophorum. Infect. Immun. 69, 5447–5455 (2001).
Yamaoka, Y. & Graham, D. Y. Helicobacter pylori virulence and cancer pathogenesis. Future Oncol. 10, 1487–1500 (2015).
Hoy, B. et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 11, 798–804 (2010).
Mallorquí-Fernández, N. et al. A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. J. Biol. Chem. 283, 2871–2882 (2008).
Kida, Y., Higashimoto, Y., Inoue, H., Shimizu, T. & Kuwano, K. A novel secreted protease from pseudomonas aeruginosa activates NF-KB through protease-activated receptors. Cell Microbiol. 10, 1491–1504 (2008).
Rosenfeld, M. E. & Campbell, L. A. Pathogens and atherosclerosis: Update on the potential contribution of multiple infectious organisms to the pathogenesis of atherosclerosis. Thromb. Haemost. 106, 858–867 (2011).
Sheikine, Y. & Hansson, G. K. Chemokines and atherosclerosis. Ann. Med. 36, 98–118 (2004).
Kholy, K., Genco, R. & Van Dyke, T. Oral infections and cardiovascular disease. Trends. Endocrinol. Metab. 26, 315–321 (2015).
Guo, L., He, X. & Shi, W. Intercellular communications in multispecies oral microbial communities. Front. Microbiol. 5, 1–13 (2014).
Filoche, S. K., Zhu, M. & Wu, C. D. In situ biofilm formation by multi-species oral bacteria under flowing and anaerobic conditions. J. Dent. Res. 83, 802–806 (2004).
Caous, J. S., Lövenklev, M., Fäldt, J. & Langton, M. Adhesion of streptococcus mitis and actinomyces oris in co-culture to machined and anodized titanium surfaces as affected by atmosphere and pH. BMC. Oral. Health. 13 doi:10.1186/1472-6831-13-4 (2013).
Periasamy, S. & Kolenbrander, P. E. Central role of the early colonizer veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of Enamel. J. Bacteriol. 192, 2965–2972 (2010).
Kolenbrander, P. E., Palmer, R. J. Jr., Periasamy, S. & Jakubovics, N. S. Oral multispecies biofilm development. Nat. Publ. Gr. 8, 471–480 (2010).
Mishra, A. et al. The actinomyces oris type 2 fimbrial shaft FimA mediates coaggregation with oral streptococci, adherence to RBC and biofilm development. Mol. Microbiol. 77, 841–854 (2010).
Jakubovics, N. S., Gill, S. R., Vickerman, M. M. & Kolenbrander, P. E. Role of hydrogen peroxide in competition and cooperation between streptococcus gordonii and actinomyces naeslundii. FEMS Microbiol. Ecology 66, 637–644 (2008).
Sharma, A., Inagaki, S., Sigurdson, W. & Kuramitsu, H. K. Synergy between tannerella forsythia and fusobacterium nucleatum in biofilm formation. Oral Microbiol. Immunol. 20, 39–42 (2005).
Kaplan, C. W., Lux, R., Haake, S. K. & Shi, W. The fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol. Microbiol. 71, 35–47 (2009).
Rosen, G., Genzler, T. & Sela, M. N. Coaggregation of treponema denticola with porphyromonas gingivalis and fusobacterium nucleatum is mediated by the major outer sheath protein of treponema denticola. FEMS Microbiol. Lett. 289, 59–66 (2008).
Rupani, D., Izano, E. A., Schreiner, H. C., Fine, D. H. & Kaplan, J. B. Aggregatibacter actinomycetemcomitans serotype f O-polysaccharide mediates coaggregation with fusobacterium nucleatum. Oral Microbiol. Immun. 23, 127–130 (2008).
Cogoni, V., Morgan-smith, A., Fenno, J. C., Jenkinson, H. F. & Dymock, D. Treponema denticola chymotrypsin-like proteinase (CTLP) integrates spirochaetes within oral microbial communities. Microbiology 158, 759–770 (2012).
Könönen, E. Pigmented prevotella species in the periodontally healthy oral cavity. FEMS Immunol. Med. Microbiol. 6, 201–205 (1993).
Rangé, H. et al. Periodontal bacteria in human carotid atherothrombosis as a potential trigger for neutrophil activation. Atherosclerosis 236, 448–455 (2014).
Kadowaki, T. et al. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J. Biochem. 128, 153–159 (2000).
Haraguchi, A., Miura, M., Fujise, O., Hamachi, T. & Nishimura, F. Porphyromonas gingivalis gingipain is involved in the detachment and aggregation of aggregatibacter actinomycetemcomitans biofilm. Mol. Oral Microbiol. 29, 131–143 (2014).
Lancy, P. Jr, Dirienzo, J. M., Appelbaum, B., Rosan, B. & Holt, S. C. Corncob formation between fusobacterium nucleatum and Streptococcus sanguis. Infect. Immun. 40, 303–309 (1983).
Lanter, B. B., Sauer, K. & Davies, D. G. Bacteria present in carotid arterial plaques are found as biofilm deposits which may contribute to enhanced risk of plaque rupture. MBio 5, e01206–e01214 (2014).
Lamont, R. J. & Kuboniwa, M. Subgingival biofilm formation. Masae Kuboniwa Richard J. Lamont. 52, 38–52 (2010).
Zarco, M. F., Vess, T. J. & Ginsburg, G. S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 18, 109–120 (2012).
Chen, H. & Jiang, W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 5, 508 (2014).
Philip D. Marsh, Thuy Do, D. B. & Deirdre A. D. Influence of saliva on the oral microbiota. Periodontology 2000 70, 82–90 (2016).
Hans, M. & Hans, V. M. Epithelial antimicrobial peptides: Guardian of the oral cavity. Int. J. Pept. 2014, 370297 (2014).
Gorr, S.-U. Antimicrobial peptides in periodontal innate defense. Front. Oral. Biol. 15, 84–98 (2012).
Rath, S. K., Mukherjee, M., Kaushik, R., Sen, S. & Kumar, M. Periodontal pathogens in atheromatous plaque. Indian J. Pathol. Microbiol. 57, 259–264 (2014).
Figuero, E. et al. Detection of periodontal bacteria in atheromatous plaque by nested polymerase chain reaction. J. Periodontol. 82, 1469–1477 (2011).
Ao, M. et al. Infection with porphyromonas gingivalis exacerbates endothelial injury in obese mice. PLoS ONE 9, e110519 (2014).
Ford, P. J. et al. Inflammation, heat shock proteins and periodontal pathogens in atherosclerosis: An immunohistologic study. Oral Microbiol. Immunol. 21, 206–211 (2006).
Taylor-Robinson, D. et al. Oro-dental bacteria in various atherosclerotic arteries. Eur. J. Clin. Microbiol. Infect. Dis. 21, 755–757 (2002).
Haraszthy, V. I., Zambon, J. J., Trevisan, M., Zeid, M. & Genco, R. J. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 71, 1554–1560 (2000).
Nakano, K. et al. Detection of cariogenic streptococcus mutans in extirpated heart valve and atheromatous plaque specimens. J. Clin. Microbiol. 44, 3313–3317 (2006).
Teles, R. & Wang, C. Y. Mechanisms involved in the association between periodontal diseases and cardiovascular disease. Oral Dis. 17, 450–461 (2011).
Gaetti-Jardim, E., Marcelino, S.L., Feitosa, A. C. R., Romito, G. A. & Avila-Campos, M. J. Quantitative detection of periodontopathic bacteria in atherosclerotic plaques from coronary arteries. J. Med. Microbiol. 58, 1568–1575 (2009).
Mahendra, J., Mahendra, L., Kurian, V. M., Jaishankar, K. & Mythilli, R. 16S rRNA-based detection of oral pathogens in coronary atherosclerotic plaque. Indian J. Dent. Res. 21, 248–252 (2010).
Ishihara, K. et al. Correlation between detection rates of periodontopathic bacterial DNA in carotid coronary stenotic artery plaque and in dental plaque samples. J. Clin. Microbiol. 42, 1313–1315 (2004).
Kuo, C. & Campbell, L. A. Is infection with chlamydia pneumoniae a causative agent in atherosclerosis?. Mol. Med. Today 4, 426–430 (1998).
Ott, S. J. et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation 113, 929–937 (2006).
Schumacher, A. et al. Does infection with chlamydia pneumoniae and/or helicobacter pylori increase the expression of endothelial cell adhesion molecules in humans?. Clin. Microbiol. Infect. 8, 654–661 (2002).
Farsak, B. et al. Detection of chlamydia pneumoniae and Helicobacter pylori DNA in human atherosclerotic plaques by PCR. J. Clin. Microbiol. 38, 4408–4411 (2000).
Cochrane, M., Pospischil, A., Walker, P., Gibbs, H. & Timms, P. Distribution of chlamydia pneumoniae DNA in atherosclerotic carotid arteries: Significance for sampling procedures. J. Clin. Microbiol. 41, 1454–1457 (2003).
Jackson, L. A. et al. Isolation of chlamydia pneumoniae from a carotid endarterectomy specimen. J. Infect. Dis. 176, 292–295 (1997).
Muhlestein, J. B. et al. Increased incidence of chlamydia species within the coronary arteries of patients with symptomatic atherosclerotic versus other forms of cardiovascular disease. J. Am. Coll. Cardiol. 27, 1555–1561 (1996).
Dobrilovic, N., Vadlamani, L., Meyer, M. & Wright, C. B. Chlamydia pneumoniae in atherosclerotic carotid artery plaques: High prevalence among heavy smokers. Am. Surg. 67, 589–593 (2001).
Oshima, T. et al. Association of helicobacter pylori infection with systemic inflammation and endothelial dysfunction in healthy male subjects. J. Am. Coll. Cardiol. 45, 1219–1222 (2005).
Mahendra, J., Mahendra, L., Nagarajan, A. & Mathew, K. Prevalence of eight putative periodontal pathogens in atherosclerotic plaque of coronary artery disease patients and comparing them with noncardiac subjects: A case-control study. Indian J. Dent. Res. 26, 189–195 (2015).
Okuda, K., Kato, T. & Ishihara, K. Involvement of periodontopathic biofilm in vascular diseases. Oral Dis. 10, 5–12 (2004).
Mahendra, J., Mahendra, L., Kurian, V. M., Jaishankar, K. & Mythilli, R. Prevalence of periodontal pathogens in coronary atherosclerotic plaque of patients undergoing coronary artery bypass graft surgery. J. Maxillof. Oral Surg. 8, 108–113 (2009).
Rafferty, B. et al. Cultivation of enterobacter hormaechei from human atherosclerotic tissue. J. Atheroscler. Thromb. 18, 72–81 (2011).
Latronico, M. et al. Periodontal disease and coronary heart disease: An epidemiological and microbiological study. New Microbiologica. 30, 221–228 (2007).
Moore, C., Addison, D., Wilson, J. M. & Zeluff, B. First case of fusobacterium necrophorum endocarditis to have presented after the 2nd decade of life. Tex. Heart Inst. J. 40, 449–452 (2013).
Samant, J. S. & Peacock, J. E. Fusobacterium necrophorum endocarditis. Case report and review of the literature. Diagn. Microbiol. Infect. Dis. 69, 192–195 (2011).
Stuart, G. & Wren, C. Endocarditis with acute mitral regurgitation caused by fusobacterium necrophorum. Pediatr. Cardiol. 13, 230–232 (1992).
Ameriso, S. F., Fridman, E. A., Leiguarda, R. C. & Sevlever, G. E. Detection of helicobacter pylori in human carotid atherosclerotic plaques. Stroke 32, 385–391 (2001).
Martínez Torres, A. & Martínez Gaensly, M. Helicobacter pylori: A new cardiovascular risk factor?. Rev. española Cardiol. 55, 652–656 (2002).
Momiyama, Y., Ohmori, R., Taniguchi, H., Nakamura, H. & Ohsuzu, F. Association of mycoplasma pneumoniae infection with coronary artery disease and its interaction with chlamydial infection. Atherosclerosis 176, 139–144 (2004).
Higuchi-Dos-Santos, M. H. et al. Chlamydia pneumoniae and mycoplasma pneumoniae in calcified nodes of stenosed aortic valves. Arq. Bras. Cardio. 84, 443–448 (2005).
Kong, H.-J., Choi, K.-K., Park, S.-H., Lee, J.-Y. & Choi, G.-W. Gene expression of human coronary artery endothelial cells in response to porphyromonas endodontalis invasion. J. Korean. Acad. Conserv. Dent. 34, 537–550 (2009).
Toyofuku, T. et al. Differential detection rate of periodontopathic bacteria in atherosclerosis. Surg. Today. 41, 1395–1400 (2011).
Curran, S. A. et al. Bacteria in the adventitia of cardiovascular disease patients with and without rheumatoid arthritis. PLoS ONE 9, e98627 (2014).
Koren, O. et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 108(Suppl), 4592–4598 (2011).
Ismail, F. et al. 16S rDNA-based metagenomic analysis of human oral plaque microbiota in patients with atherosclerosis and healthy controls. Indian J. Med. Microbiol. 30, 462–466 (2012).
DiFranco, K. M. et al. Leukotoxin (Leukothera®) targets active leukocyte function antigen-1 (LFA-1) protein and triggers a lysosomal mediated cell death pathway. J. Biol. Chem. 287, 17618–17627 (2012).
Ando-Suguimoto, E. S. et al. The cytolethal distending toxin of aggregatibacter actinomycetemcomitans inhibits macrophage phagocytosis and subverts cytokine production. Cytokine 66, 46–53 (2014).
Jorgensen, I. et al. The chlamydia protease CPAF regulates host and bacterial proteins to maintain pathogen vacuole integrity and promote virulence. Cell Host Microbe 10, 21–32 (2011).
Shaw, A. C. et al. Characterization of a secreted chlamydia protease. Cell Microbiol. 4, 411–424 (2002).
Apolônio, A. C. et al. Purification and partial characterization of a bacteriocin produced by eikenella corrodens. J. Appl. Microbiol. 104, 508–514 (2008).
Allaker, R. P., Young, K. A. & Hardie, J. M. Production of hydrolytic enzymes by oral isolates of eikenella corrodens. FEMS Microbiol. Lett. 123, 69–74 (1994).
Doron, L. et al. Identification and characterization of fusolisin, the fusobacterium nucleatum autotransporter serine protease. PLoS ONE 9, e111329 (2014).
Kusters, J. G., Van Vliet, A. H. M. & Kuipers, E. J. Pathogenesis of helicobacter pylori infection. Clin. Microbiol. Rev. 19, 449–490 (2006).
Palframan, S. L., Kwok, T. & Gabriel, K. Vacuolating cytotoxin A (VacA), a key toxin for helicobacter pylori pathogenesis. Front. Cell Infect. Microbiol. 2, 1–9 (2012).
Fu, H.-W. Helicobacter pylori neutrophil-activating protein: From molecular pathogenesis to clinical applications. World J. Gastroenterol. 20, 5294–5301 (2014).
Yeh, Y.-C., Cheng, H.-C., Yang, H.-B., Chang, W.-L. & Sheu, B. Pylori CagL-Y58/E59 prime higher integrin α5β1 in adverse pH condition to enhance hypochlorhydria vicious cycle for gastric carcinogenesis. PLoS ONE 8, e72735 (2013).
Tang, C.-L. Helicobacter pylori tumor necrosis factor-α inducing protein promotes cytokine expression via nuclear factor-κB. World J. Gastroenterol. 19, 399 (2013).
Li, J., Meng, F. L., He, L. H. & Zhang, J. Z. Secreted protein HP1286 of helicobacter pylori strain 26695 induces apoptosis of AGS cells. Biomed. Environ. Sci. 25, 614–619 (2012).
Lester, J. et al. Characterization of helicobacter pylori HP0231 (DsbK): Role in disulfide bond formation, redox homeostasis and production of helicobacter cystein-rich protein HcpE. Mol. Microbiol. 96, 110–133 (2015).
Rosen, G., Shoshani, M., Naor, R. & Sela, M. N. The purificat0ion and characterization of an 88-kDa porphyromonas endodontalis 35406 protease. Oral Microbiol. Immunol. 16, 326–331 (2001).
Yoshimura, F., Takahashi, K., Nodasaka, Y. & Suzuki, T. Purification and characterization of a novel type of fimbriae from the oral anaerobe bacteroides gingivalis. J. Bacteriol. 160, 949–957 (1984).
Nelson, D. et al. pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: Purification, characterization, and cloning of the gene encoding this enzyme. SAGE J. 80, 371–377 (2001).
Jefferies, J. et al. Identification of a secreted cholesterol-dependent cytolysin (mitilysin) from streptococcus mitis. J. Bacteriol. 189, 627–632 (2007).
Jarosz, L. M., Deng, D. M., van der Mei, H. C., Crielaard, W. & Krom, B. P. Streptococcus mutans competence-stimulating peptide inhibits candida albicans hypha formation. Eukaryot. Cell. 8, 1658–1664 (2009).
Burne, R. A. & Penders, J. E. Characterization of the streptococcus mutans GS-5 fruA gene encoding exo-beta-D-fructosidase. Infect. Immun. 60, 4621–4632 (1992).
Cerdà-Costa, N. et al. The structure of the catalytic domain of tannerella forsythia karilysin reveals it is a bacterial xenologue of animal matrix metalloproteinases. Mol. Microbiol. 79, 119–132 (2011).
Ksiazek, M. et al. KLIKK proteases of tannerella forsythia: putative virulence factors with a unique domain structure. Front. Microbiol. 6, 312 (2015).
Ishihara, K. et al. Dentipain, a streptococcus pyogenes IdeS protease homolog, is a novel virulence factor of treponema denticola. Biol. Chem. 391, 1047–1055 (2010).
Acknowledgements
We thank CB Indus for constant encouragement. This work was supported by DBT OSRP grant and JC Bose fellowship to AS.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chhibber-Goel, J., Singhal, V., Bhowmik, D. et al. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. npj Biofilms Microbiomes 2, 7 (2016). https://doi.org/10.1038/s41522-016-0009-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-016-0009-7
This article is cited by
-
Exploring the presence of oral bacteria in non-oral sites of patients with cardiovascular diseases using whole metagenomic data
Scientific Reports (2024)
-
Multi-site microbiota alteration is a hallmark of kidney stone formation
Microbiome (2023)
-
The oral microbiome in the pathophysiology of cardiovascular disease
Nature Reviews Cardiology (2023)
-
Epigenetics and the role of nutraceuticals in health and disease
Environmental Science and Pollution Research (2023)
-
The Gut Microbiome, Microbial Metabolites, and Cardiovascular Disease in People Living with HIV
Current HIV/AIDS Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.