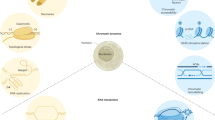
Abstract
The plant DNA damage response (DDR) pathway safeguards genomic integrity by rapid recognition and repair of DNA lesions that, if unrepaired, may cause genome instability. Most frequently, DNA repair goes hand in hand with a transient cell cycle arrest, which allows cells to repair the DNA lesions before engaging in a mitotic event, but consequently also affects plant growth and yield. Through the identification of DDR proteins and cell cycle regulators that react to DNA double-strand breaks or replication defects, it has become clear that these proteins and regulators form highly interconnected networks. These networks operate at both the transcriptional and post-transcriptional levels and include liquid–liquid phase separation and epigenetic mechanisms. Strikingly, whereas the upstream DDR sensors and signalling components are well conserved across eukaryotes, some of the more downstream effectors are diverged in plants, probably to suit unique lifestyle features. Additionally, DDR components display functional diversity across ancient plant species, dicots and monocots. The observed resistance of DDR mutants towards aluminium toxicity, phosphate limitation and seed ageing indicates that gaining knowledge about the plant DDR may offer solutions to combat the effects of climate change and the associated risk for food security.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pedroza-Garcia, J. A., Xiang, Y. & De Veylder, L. Cell cycle checkpoint control in response to DNA damage by environmental stresses. Plant J. 109, 490–507 (2022).
Langerak, P., Mejia-Ramirez, E., Limbo, O. & Russell, P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271 (2011).
Lee, J. H. & Paull, T. T. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science 308, 551–554 (2005).
Aklilu, B. B., Soderquist, R. S. & Culligan, K. M. Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res. 42, 3104–3118 (2014).
Yates, L. A. et al. A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA. Nat. Commun. 9, 5447 (2018).
Eschbach, V. & Kobbe, D. Different replication protein A complexes of Arabidopsis thaliana have different DNA-binding properties as a function of heterotrimer composition. Plant Cell Physiol. 55, 1460–1472 (2014).
Zou, L. & Elledge, S. J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300, 1542–1548 (2003).
Sweeney, P. R., Britt, A. B. & Culligan, K. M. The Arabidopsis ATRIP ortholog is required for a programmed response to replication inhibitors. Plant J. 60, 518–526 (2009).
Cortez, D., Guntuku, S., Qin, J. & Elledge, S. J. ATR and ATRIP: partners in checkpoint signaling. Science 294, 1713–1716 (2001).
Namiki, Y. & Zou, L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc. Natl Acad. Sci. USA 103, 580–585 (2006).
Kumagai, A., Lee, J., Yoo, H. Y. & Dunphy, W. G. TopBP1 activates the ATR–ATRIP complex. Cell 124, 943–955 (2006).
Heitzeberg, F. et al. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 38, 954–968 (2004).
Zou, L., Liu, D. & Elledge, S. J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl Acad. Sci. USA 100, 13827–13832 (2003).
Banin, S. et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677 (1998).
Canman, C. E. et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 (1998).
Tibbetts, R. S. et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13, 152–157 (1999).
Preuss, S. B. & Britt, A. B. A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 164, 323–334 (2003).
Yoshiyama, K. O., Conklin, P. A., Huefner, N. D. & Britt, A. B. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl Acad. Sci. USA 106, 12843–12848 (2009).
Sjogren, C. A., Bolaris, S. C. & Larsen, P. B. Aluminum-dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR-, ALT2-, and SOG1-regulated transcriptional response. Plant Cell 27, 2501–2515 (2015).
Yoshiyama, K. O. et al. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 14, 817–822 (2013).
Bourbousse, C., Vegesna, N. & Law, J. A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl Acad. Sci. USA 115, E12453–E12462 (2018).
Ogita, N. et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 94, 439–453 (2018).
Vandepoele, K. et al. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139, 316–328 (2005).
Nisa, M. et al. Distinctive and complementary roles of E2F transcription factors during plant replication stress responses. Mol. Plant 16, 1269–1282 (2023).
Waterworth, W. M. et al. Phosphoproteomic analysis reveals plant DNA damage signalling pathways with a functional role for histone H2AX phosphorylation in plant growth under genotoxic stress. Plant J. 100, 1007–1021 (2019).
Fan, T. et al. Arabidopsis γ-H2A.X-INTERACTING PROTEIN participates in DNA damage response and safeguards chromatin stability. Nat. Commun. 13, 7942 (2022).
Vladejic, J. et al. Analysis of BRCT5 domain-containing proteins reveals a new component of DNA damage repair in Arabidopsis. Front. Plant Sci. 13, 1023358 (2022).
Yu, X., Chini, C. C. S., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003).
Lorković, Z. J., Klingenbrunner, M., Cho, C. H. & Berger, F. Identification of plants functional counterparts of the metazoan Mediator of DNA Damage Checkpoint 1. Preprint at bioRxiv https://doi.org/10.1101/2023.05.19.541430 (2023).
Biedermann, S. et al. The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J. 36, 1279–1297 (2017).
Yu, C. et al. The multi-BRCT domain protein DDRM2 promotes the recruitment of RAD51 to DNA damage sites to facilitate homologous recombination. N. Phytol. 238, 1073–1084 (2023).
Weimer, A. K. et al. The plant-specific CDKB1–CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 35, 2068–2086 (2016).
Horvath, B. M. et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 36, 1261–1278 (2017).
Kurzbauer, M.-T. et al. ATM controls meiotic DNA double-strand break formation and recombination and affects synaptonemal complex organization in plants. Plant Cell 33, 1633–1656 (2021).
Meschichi, A. et al. The plant-specific DDR factor SOG1 increases chromatin mobility in response to DNA damage. EMBO Rep. 23, e54736 (2022).
Yin, C., Sun, A., Guo, T., Mao, X. & Fang, Y. Arabidopsis lamin-like proteins CRWN1 and CRWN2 interact with SUPPRESSOR OF NPR1-1 INDUCIBLE 1 and RAD51D to prevent DNA damage. Plant Cell 35, 3345–3362 (2023).
Watanabe, K. et al. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21, 2688–2699 (2009).
Wei, W. et al. A role for small RNAs in DNA double-strand break repair. Cell 149, 101–112 (2012).
Jiang, J. et al. A diRNA–protein scaffold module mediates SMC5/6 recruitment in plant DNA repair. Plant Cell 34, 3899–3914 (2022).
Lai, J. et al. The transcriptional coactivator ADA2b recruits a structural maintenance protein to double-strand breaks during DNA repair in plants. Plant Physiol. 176, 2613–2622 (2018).
Jiang, J. et al. A SWI/SNF subunit regulates chromosomal dissociation of structural maintenance complex 5 during DNA repair in plant cells. Proc. Natl Acad. Sci. USA 116, 15288–15296 (2019).
Li, C., Guo, Y., Wang, L. & Yan, S. The SMC5/6 complex recruits the PAF1 complex to facilitate DNA double-strand break repair in Arabidopsis. EMBO J. 42, e112756 (2023).
Guo, T. et al. Photoexcited cryptochromes interact with ADA2b and SMC5 to promote the repair of DNA double-strand breaks in Arabidopsis. Nat. Plants 9, 1280–1290 (2023).
Liu, M. et al. IDN2 interacts with RPA and facilitates DNA double-strand break repair by homologous recombination in Arabidopsis. Plant Cell 29, 589–599 (2017).
Wang, L. et al. The ATR–WEE1 kinase module inhibits the MAC complex to regulate replication stress response. Nucleic Acids Res. 49, 1411–1425 (2021).
Dvorak Tomastikova, E. et al. SMC5/6 complex-mediated SUMOylation stimulates DNA–protein cross-link repair in Arabidopsis. Plant Cell 35, 1532–1547 (2023).
Gentric, N., Genschik, P. & Noir, S. Connections between the cell cycle and the DNA damage response in plants. Int. J. Mol. Sci. 22, 9558 (2021).
De Schutter, K. et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19, 211–225 (2007).
Culligan, K., Tissier, A. & Britt, A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104 (2004).
Lundgren, K. et al. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64, 1111–1122 (1991).
Watanabe, N., Broome, M. & Hunter, T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 14, 1878–1891 (1995).
Dissmeyer, N. et al. Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21, 3641–3654 (2009).
Kim, H. J. et al. Control of plant germline proliferation by SCFFBL17 degradation of cell cycle inhibitors. Nature 455, 1134–1137 (2008).
Gusti, A. et al. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS ONE 4, e4780 (2009).
Pan, T. et al. A novel WEE1 pathway for replication stress responses. Nat. Plants 7, 209–218 (2021).
Pan, T., Gao, S., Cui, X., Wang, L. & Yan, S. APC/CCDC20 targets SCFFBL17 to activate replication stress responses in Arabidopsis. Plant Cell 35, 910–923 (2023).
Gentric, N. et al. The F-box-like protein FBL17 is a regulator of DNA-damage response and colocalizes with RETINOBLASTOMA RELATED1 at DNA lesion sites. Plant Physiol. 183, 1295–1305 (2020).
Bao, W. et al. Protein kinase ATR inhibits E3 ubiquitin ligase CRL4PRL1 to stabilize ribonucleotide reductase in response to replication stress. Cell Rep. 42, 112685 (2023).
Kalhorzadeh, P. et al. Arabidopsis thaliana RNase H2 deficiency counteracts the needs for the WEE1 checkpoint kinase but triggers genome instability. Plant Cell 26, 3680–3692 (2014).
Eekhout, T. et al. G2/M-checkpoint activation in fasciata1 rescues an aberrant S-phase checkpoint but causes genome instability. Plant Physiol. 186, 1893–1907 (2021).
Eekhout, T., Pedroza-Garcia, J. A., Kalhorzadeh, P., De Jaeger, G. & De Veylder, L. A mutation in DNA polymerase α rescues WEE1KO sensitivity to HU. Int. J. Mol. Sci. 22, 9409 (2021).
Hu, Z., Cools, T., Kalhorzadeh, P., Heyman, J. & De Veylder, L. Deficiency of the Arabidopsis helicase RTEL1 triggers a SOG1-dependent replication checkpoint in response to DNA cross-links. Plant Cell 27, 149–161 (2015).
Wei, P. et al. Arabidopsis casein kinase 2 triggers stem cell exhaustion under Al toxicity and phosphate deficiency through activating the DNA damage response pathway. Plant Cell 33, 1361–1380 (2021).
Hamasaki, H. et al. SnRK1 kinase and the NAC transcription factor SOG1 are components of a novel signaling pathway mediating the low energy response triggered by ATP depletion. Front. Plant Sci. 10, 503 (2019).
Chen, H. et al. The ATR–WEE1 kinase module promotes SUPPRESSOR OF GAMMA RESPONSE 1 translation to activate replication stress responses. Plant Cell 35, 3021–3034 (2023).
Wang, X. et al. A plant-specific module for homologous recombination repair. Proc. Natl Acad. Sci. USA 119, e2202970119 (2022).
Cools, T. et al. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23, 1435–1448 (2011).
Yi, D. et al. The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 26, 296–309 (2014).
Takahashi, N. et al. A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis. eLife 8, e43944 (2019).
Chen, P. et al. Arabidopsis R1R2R3-Myb proteins are essential for inhibiting cell division in response to DNA damage. Nat. Commun. 8, 635 (2017).
Lammens, T., Li, J., Leone, G. & De Veylder, L. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol. 19, 111–118 (2009).
Lang, L. et al. The DREAM complex represses growth in response to DNA damage in Arabidopsis. Life Sci. Alliance 4, e202101141 (2021).
Zaragoza, J. Z., Klap, K., Heidstra, R., Zhou, W. & Scheres, B. The dual role of the RETINOBLASTOMA-RELATED protein in the DNA damage response is coordinated by the interaction with LXCXE-containing proteins. Plant J., https://doi.org/10.1111/tpj.16665 (2024).
Ning, Y.-Q. et al. DREAM complex suppresses DNA methylation maintenance genes and precludes DNA hypermethylation. Nat. Plants 6, 942–956 (2020).
Wang, Y. et al. The Arabidopsis DREAM complex antagonizes WDR5A to modulate histone H3K4me2/3 deposition for a subset of genome repression. Proc. Natl Acad. Sci. USA 119, e2206075119 (2022).
Wang, L., Chen, H., Wang, C., Hu, Z. & Yan, S. Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proc. Natl Acad. Sci. USA 115, E3837–E3845 (2018).
Fulcher, N. & Sablowski, R. Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl Acad. Sci. USA 106, 20984–20988 (2009).
Li, G., Deng, L., Huang, N. & Sun, F. The biological roles of lncRNAs and future prospects in clinical application. Diseases 9, 8 (2021).
Wang, Z., Schwacke, R. & Kunze, R. DNA damage-induced transcription of transposable elements and long non-coding RNAs in Arabidopsis is rare and ATM-dependent. Mol. Plant 9, 1142–1155 (2016).
Durut, N. et al. Long non-coding RNAs contribute to DNA damage resistance in Arabidopsis thaliana. Genetics 225, iyad135 (2023).
Herbst, J., Nagy, S. H., Vercauteren, I., De Veylder, L. & Kunze, R. The long non-coding RNA LINDA restrains cellular collapse following DNA damage in Arabidopsis thaliana. Plant J. 116, 1370–1384 (2023).
Mortimore, G. E., Lardeux, B. R. & Adams, C. E. Regulation of microautophagy and basal protein turnover in rat liver: effects of short-term starvation. J. Biol. Chem. 263, 2506–2512 (1988).
Juretschke, T. & Beli, P. Causes and consequences of DNA damage-induced autophagy. Matrix Biol. 100–101, 39–53 (2021).
Zhang, Y. et al. Autophagy-related proteins in genome stability: autophagy-dependent and independent actions. Int. J. Biol. Sci. 18, 5329–5344 (2022).
Chen, P. et al. KNO1-mediated autophagic degradation of the Bloom syndrome complex component RMI1 promotes homologous recombination. EMBO J. 42, e111980 (2023).
Martens, M., Horres, R., Wendeler, E. & Reiss, B. The importance of ATM and ATR in Physcomitrella patens DNA damage repair, development, and gene targeting. Genes 11, 752 (2020).
Gu, N. et al. DNA damage triggers reprogramming of differentiated cells into stem cells in Physcomitrella. Nat. Plants 6, 1098–1105 (2020).
Culligan, K. M., Robertson, C. E., Foreman, J., Doerner, P. & Britt, A. B. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48, 947–961 (2006).
Yoshiyama, K. O. SOG1: a master regulator of the DNA damage response in plants. Genes Genet. Syst. 90, 209–216 (2016).
Yoshiyama, K. O., Kimura, S., Maki, H., Britt, A. B. & Umeda, M. The role of SOG1, a plant-specific transcriptional regulator, in the DNA damage response. Plant Signal. Behav. 9, e28889 (2014).
Goffová, I. et al. Roles of RAD51 and RTEL1 in telomere and rDNA stability in Physcomitrella patens. Plant J. 98, 1090–1105 (2019).
Sakamoto, A. N. et al. SOG1, a plant-specific master regulator of DNA damage responses, originated from nonvascular land plants. Plant Direct 5, e370 (2021).
Pedroza-Garcia, J. A. et al. Maize ATR safeguards genome stability during kernel development to prevent early endosperm endocycle onset and cell death. Plant Cell 33, 2662–2684 (2021).
Szurman-Zubrzycka, M. et al. ATR, a DNA damage signaling kinase, is involved in aluminum response in barley. Front. Plant Sci. 10, 1299 (2019).
Haberer, G. et al. Structure and architecture of the maize genome. Plant Physiol. 139, 1612–1624 (2005).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433 (2017).
Nishizawa-Yokoi, A. et al. SUPPRESSOR OF GAMMA RESPONSE 1 plays rice-specific roles in DNA damage response and repair. Plant Physiol. 191, 1288–1304 (2023).
Tripathi, D., Oldenburg, D. J. & Bendich, A. J. Oxidative and glycation damage to mitochondrial DNA and plastid DNA during plant development. Antioxidants 12, 891 (2023).
Tripathi, D., Nam, A., Oldenburg, D. J. & Bendich, A. J. Reactive oxygen species, antioxidant agents, and DNA damage in developing maize mitochondria and plastids. Front. Plant Sci. 11, 596 (2020).
Kumar, R. A., Oldenburg, D. J. & Bendich, A. J. Changes in DNA damage, molecular integrity, and copy number for plastid DNA and mitochondrial DNA during maize development. J. Exp. Bot. 65, 6425–6439 (2014).
Maréchal, A. et al. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl Acad. Sci. USA 106, 14693–14698 (2009).
Zampini, E., Lepage, E., Tremblay-Belzile, S., Truche, S. & Brisson, N. Organelle DNA rearrangement mapping reveals U-turn-like inversions as a major source of genomic instability in Arabidopsis and humans. Genome Res. 25, 645–654 (2015).
Wang, W. et al. RNase H1C collaborates with ssDNA binding proteins WHY1/3 and recombinase RecA1 to fulfill the DNA damage repair in Arabidopsis chloroplasts. Nucleic Acids Res. 49, 6771–6787 (2021).
Duan, S., Hu, L., Dong, B., Jin, H.-L. & Wang, H.-B. Signaling from plastid genome stability modulates endoreplication and cell cycle during plant development. Cell Rep. 32, 108019 (2020).
Hudik, E. et al. Chloroplast dysfunction causes multiple defects in cell cycle progression in the Arabidopsis crumpled leaf mutant. Plant Physiol. 166, 152–167 (2014).
Jin, H.-L. et al. Dual roles for CND1 in maintenance of nuclear and chloroplast genome stability in plants. Cell Rep. 42, 112268 (2023).
Han, S.-H., Park, Y.-J. & Park, C.-M. HOS1 activates DNA repair systems to enhance plant thermotolerance. Nat. Plants 6, 1439–1446 (2020).
Zhuang, K. et al. WHIRLY1 regulates HSP21.5A expression to promote thermotolerance in tomato. Plant Cell Physiol. 61, 169–177 (2020).
Babbar, R., Karpinska, B., Grover, A. & Foyer, C. H. Heat-induced oxidation of the nuclei and cytosol. Front. Plant Sci. 11, 617779 (2020).
Velichko, A. K., Petrova, N. V., Kantidze, O. L. & Razin, S. V. Dual effect of heat shock on DNA replication and genome integrity. Mol. Biol. Cell 23, 3450–3460 (2012).
Lloyd, A., Morgan, C., Franklin, F. C. H. & Bomblies, K. Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 208, 1409–1420 (2018).
Ning, Y. et al. Heat stress interferes with formation of double-strand breaks and homolog synapsis. Plant Physiol. 185, 1783–1797 (2021).
Lee, J. R. et al. Dynamic interactions of Arabidopsis TEN1: stabilizing telomeres in response to heat stress. Plant Cell 28, 2212–2224 (2016).
Belfield, E. J. et al. Thermal stress accelerates Arabidopsis thaliana mutation rate. Genome Res. 31, 40–50 (2021).
Perkins-Kirkpatrick, S. E. & Lewis, S. C. Increasing trends in regional heatwaves. Nat. Commun. 11, 3357 (2020).
Kobbe, D., Blanck, S., Demand, K., Focke, M. & Puchta, H. AtRECQ2, a RecQ helicase homologue from Arabidopsis thaliana, is able to disrupt various recombinogenic DNA structures in vitro. Plant J. 55, 397–405 (2008).
Zhao, J. et al. ATM-mediated double-strand break repair is required for meiotic genome stability at high temperature. Plant J. 114, 403–423 (2023).
Zhao, L. et al. Transcriptional regulation of cell cycle genes in response to abiotic stresses correlates with dynamic changes in histone modifications in maize. PLoS ONE 9, e106070 (2014).
Fenollosa, E., Jene, L. & Munne-Bosch, S. A rapid and sensitive method to assess seed longevity through accelerated aging in an invasive plant species. Plant Methods 16, 64 (2020).
De Vitis, M. et al. Seed storage: maintaining seed viability and vigor for restoration use. Restor. Ecol. 28, S249–S255 (2020).
Waterworth, W. M., Latham, R., Wang, D., Alsharif, M. & West, C. E. Seed DNA damage responses promote germination and growth in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 119, e2202172119 (2022).
Li, Z. et al. ATM suppresses leaf senescence triggered by DNA double-strand break through epigenetic control of senescence-associated genes in Arabidopsis. N. Phytol. 227, 473–484 (2020).
Waterworth, W. M., Footitt, S., Bray, C. M., Finch-Savage, W. E. & West, C. E. DNA damage checkpoint kinase ATM regulates germination and maintains genome stability in seeds. Proc. Natl Acad. Sci. USA 113, 9647–9652 (2016).
Müller, J. et al. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 33, 216–230 (2015).
Zheng, Z., Wang, Z., Wang, X. & Liu, D. Blue light-triggered chemical reactions underlie phosphate deficiency-induced inhibition of root elongation of Arabidopsis seedlings grown in petri dishes. Mol. Plant 12, 1515–1523 (2019).
Reyt, G., Boudouf, S., Boucherez, J., Gaymard, F. & Briat, J.-F. Iron- and ferritin-dependent reactive oxygen species distribution: impact on Arabidopsis root system architecture. Mol. Plant 8, 439–453 (2015).
Latha, K. S., Anitha, S., Rao, K. S. J. & Viswamitra, M. A. Molecular understanding of aluminum-induced topological changes in CCG12 triplet repeats: relevance to neurological disorders. Biochim. Biophys. Acta Mol. Basis Dis. 1588, 56–64 (2002).
Nezames, C. D., Sjogren, C. A., Barajas, J. F. & Larsen, P. B. The Arabidopsis cell cycle checkpoint regulators TANMEI/ALT2 and ATR mediate the active process of aluminum-dependent root growth inhibition. Plant Cell 24, 608–621 (2012).
Zhang, Y. et al. The cell cycle checkpoint regulator ATR is required for internal aluminum toxicity-mediated root growth inhibition in Arabidopsis. Front. Plant Sci. 9, 118 (2018).
Liu, D., Jiang, W., Zhang, L. & Li, L. Effects of boron ions on root growth and cell division of broadbean (Vicia faba L.). Isr. J. Plant Sci. 48, 47–51 (2000).
Reid, R. J., Hayes, J. E., Post, A., Stangoulis, J. C. R. & Graham, R. D. A critical analysis of the causes of boron toxicity in plants. Plant Cell Environ. 27, 1405–1414 (2004).
Cervilla, L. M., Blasco, B., Ríos, J. J., Romero, L. & Ruiz, J. M. Oxidative stress and antioxidants in tomato (Solanum lycopersicum) plants subjected to boron toxicity. Ann. Bot. 100, 747–756 (2007).
Sakamoto, T. et al. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell 23, 3533–3546 (2011).
Sakamoto, T. et al. Proteasomal degradation of BRAHMA promotes boron tolerance in Arabidopsis. Nat. Commun. 9, 5285 (2018).
Sotta, N. et al. NAC103 mutation alleviates DNA damage in an Arabidopsis thaliana mutant sensitive to excess boron. Front. Plant Sci. 14, 1099816 (2023).
Hendrix, S. et al. Suppressor of gamma response 1 modulates the DNA damage response and oxidative stress response in leaves of cadmium-exposed Arabidopsis thaliana. Front. Plant Sci. 11, 366 (2020).
Hendrix, S. et al. Cell cycle regulation in different leaves of Arabidopsis thaliana plants grown under control and cadmium-exposed conditions. Environ. Exp. Bot. 155, 441–452 (2018).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Acknowledgements
We thank A. Bleys, M. Dubois and J. Heyman for critical reading and editing of the manuscript. This work was supported by grants from the Research Foundation Flanders (projects G011420N and G0A6J24N).
Author information
Authors and Affiliations
Contributions
L.D.V. led the project. J.H., Q.-Q.L. and L.D.V. wrote the manuscript. J.H. and Q.-Q.L. prepared the figures. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Shunping Yan, Pascal Genschik and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Herbst, J., Li, QQ. & De Veylder, L. Mechanistic insights into DNA damage recognition and checkpoint control in plants. Nat. Plants 10, 539–550 (2024). https://doi.org/10.1038/s41477-024-01652-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-024-01652-9