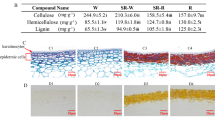
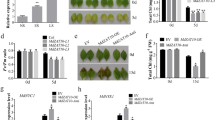
Abstract
Cuticular waxes play important roles in plant development and the interaction between plants and their environment. Researches on wax biosynthetic pathways have been reported in several plant species. Also, wax formation is closely related to environmental condition. However, the regulatory mechanism between wax and environmental factors, especially essential mineral elements, is less studied. Here we found that nitrogen (N) played a negative role in the regulation of wax synthesis in apple. We therefore analysed wax content, composition and crystals in BTB-TAZ domain protein 2 (MdBT2) overexpressing and antisense transgenic apple seedlings and found that MdBT2 could downregulate wax biosynthesis. Furthermore, R2R3-MYB transcription factor 16-like protein (MdMYB106) interacted with MdBT2, and MdBT2 mediated its ubiquitination and degradation through the 26S proteasome pathway. Finally, HXXXD-type acyl-transferase ECERIFERUM 2-like1 (MdCER2L1) was confirmed as a downstream target gene of MdMYB106. Our findings reveal an N-mediated apple wax biosynthesis pathway and lay a foundation for further study of the environmental factors associated with wax regulatory networks in apple.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The authors confirm that all experimental data are available and accessible via the main text and/or the supplemental data. The accession codes are listed as follows: MdBT2: MD06G1161300; MdMYB106: MD09G1054000; MdCER2: MD15G1386800; MdCER2L1: MD15G1386300; AtCER2: AT4G24510; MdCER6: MD13G1042400. Additional data related to this study are available from the corresponding author upon request. All biological materials used in this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Kaur, G. et al. Adaptation to early-season soil waterlogging using different nitrogen fertilizer practices and corn hybrids. Agronomy 10, 378 (2020).
Chen, Z. et al. Expression analysis of nitrogen metabolism-related genes reveals differences in adaptation to low-nitrogen stress between two different barley cultivars at seedling stage. Int. J. Genomics 2018, 8152860 (2018).
Sharma, S., Singh, P., Kaur, S. & Singh, Y. Fertilizer-N application and rice straw incorporation impacts on crop yields, potassium use efficiency and potassium fractions in a rice–wheat cropping system. Commun. Soil Sci. Plant Anal. 53, 793–813 (2022).
Yasuda, S. et al. Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14-3-3 proteins. J. Biol. Chem. 289, 15179–15193 (2014).
Ge, L. et al. SiMYB3 in foxtail millet (Setaria italica) confers tolerance to low-nitrogen stress by regulating root growth in transgenic plants. Int. J. Mol. Sci. 20, 5741 (2019).
Drazeta, L. et al. Causes and effects of changes in xylem functionality in apple fruit. Ann. Bot. 93, 275–282 (2004).
Dong, B. & Guo, L. An efficient gene disruption method for the woody plant pathogen Botryosphaeria dothidea. BMC Biotechnol. 20, 14 (2020).
De Miccolis Angelini, R. M. et al. De novo assembly and comparative transcriptome analysis of Monilinia fructicola, Monilinia laxa and Monilinia fructigena, the causal agents of brown rot on stone fruits. BMC Genomics 19, 436 (2018).
Sun, M. et al. Endophytic bacterium Serratia plymuthica from Chinese leek suppressed apple ring rot on postharvest apple fruit. Front. Microbiol. 12, 802887 (2022).
Chou, C. et al. Expression of five endopolygalacturonase genes and demonstration that MfPG1 overexpression diminishes virulence in the brown rot pathogen Monilinia fructicola. PLoS ONE 10, e132012 (2015).
El-Garhy, H. A. S. et al. Comparative analyses of four chemicals used to control black mold disease in tomato and its effects on defense signaling pathways, productivity and quality traits. Plants 9, 808 (2020).
Samuels, L., Kunst, L. & Jetter, R. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707 (2008).
Lü, S. et al. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59, 553–564 (2009).
Kim, H. et al. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53, 1391–1403 (2012).
Zheng, J. et al. Co-expression analysis aids in the identification of genes in the cuticular wax pathway in maize. Plant J. 97, 530–542 (2019).
Pascal, S. et al. Arabidopsis CER1-LIKE1 functions in a cuticular very-long-chain alkane-forming complex. Plant Physiol. 179, 415–432 (2019).
Kim, H., Go, Y. S. & Suh, M. C. DEWAX2 transcription factor negatively regulates cuticular wax biosynthesis in Arabidopsis leaves. Plant Cell Physiol. 59, 966–977 (2018).
Sajeevan, R. S. et al. Expression of Arabidopsis SHN1 in Indian mulberry (Morus indica L.) increases leaf surface wax content and reduces post-harvest water loss. Front. Plant Sci. 8, 418 (2017).
Domergue, F. et al. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 153, 1539–1554 (2010).
Zhang, Y., You, C., Li, Y. & Hao, Y. Advances in biosynthesis, regulation, and function of apple cuticular wax. Front. Plant Sci. 11, 1165 (2020).
Zhang, Y. et al. Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 249, 1627–1643 (2019).
Yang, H. et al. CitWRKY28 and CitNAC029 promote the synthesis of cuticular wax by activating CitKCS gene expression in citrus fruit. Plant Cell Rep. 41, 905–920 (2022).
Zhu, X. et al. Comparative study of volatile compounds in the fruit of two banana cultivars at different ripening stages. Molecules 23, 2456 (2018).
Yun, Z. et al. Comparative transcriptomics and proteomics analysis of citrus fruit, to improve understanding of the effect of low temperature on maintaining fruit quality during lengthy post-harvest storage. J. Exp. Bot. 63, 2873–2893 (2012).
Zhong, M. et al. MdCER2 conferred to wax accumulation and increased drought tolerance in plants. Plant Physiol. Biochem. 149, 277–285 (2020).
Haslam, T. M., Mañas-Fernández, A., Zhao, L. & Kunst, L. Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 160, 1164–1174 (2012).
Haslam, T., Gerelle, W., Graham, S. & Kunst, L. The unique role of the ECERIFERUM2-LIKE clade of the BAHD acyltransferase superfamily in cuticular wax metabolism. Plants 6, 23 (2017).
Qi, C. et al. Isolation and functional identification of an apple MdCER1 gene. Plant Cell Tissue Organ Cult. 136, 1–13 (2019).
Lashbrooke, J. et al. The tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol. 169, 2553–2571 (2015).
Lashbrooke, J. et al. MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. Plant Cell 28, 2097–2116 (2016).
Jiang, L. et al. MdGH3.6 is targeted by MdMYB94 and plays a negative role in apple water-deficit stress tolerance. Plant J. 109, 1271–1289 (2022).
Lee, S. B., Kim, H. U. & Suh, M. C. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol. 57, 2300–2311 (2016).
Zhang, Y. et al. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 19, 362 (2019).
Feng, X., An, Y., Zheng, J., Sun, M. & Wang, L. Proteomics and SSH analyses of ALA-promoted fruit coloration and evidence for the involvement of a MADS-box gene, MdMADS1. Front. Plant Sci. 7, 1615 (2016).
Zhao, Q. et al. Ubiquitination-related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol. 172, 1973–1988 (2016).
Wang, X. et al. The nitrate-responsive protein MdBT2 regulates anthocyanin biosynthesis by interacting with the MdMYB1 transcription factor. Plant Physiol. 178, 890–906 (2018).
An, J. et al. Apple BT2 protein negatively regulates jasmonic acid‐triggered leaf senescence by modulating the stability of MYC2 and JAZ2. Plant Cell Environ. 44, 216–233 (2020).
An, J. P., Wang, X. F. & Hao, Y. BTB/TAZ protein MdBT2 integrates multiple hormonal and environmental signals to regulate anthocyanin biosynthesis in apple. J. Integr. Plant Biol. 62, 1643–1646 (2020).
An, J. et al. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 17, 2231–2233 (2019).
An, J. et al. MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 222, 735–751 (2019).
An, J. et al. MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol. 224, 380–395 (2019).
Xu, J. et al. Dynamic transcription profiles of “Qinguan” apple (Malus × domestica) leaves in response to Marssonina coronaria inoculation. Front. Plant Sci. 6, 842 (2015).
Ding, N. et al. Effects of crop load on distribution and utilization of 13C and 15N and fruit quality for dwarf apple trees. Sci. Rep. 7, 14172 (2017).
Xu, X. et al. Effects of potassium levels on plant growth, accumulation and distribution of carbon, and nitrate metabolism in apple dwarf rootstock seedlings. Front. Plant Sci. 11, 904 (2020).
Wang, F. et al. Nitrification inhibitor 3,4-dimethylpyrazole phosphate application during the later stage of apple fruit expansion regulates soil mineral nitrogen and tree carbon–nitrogen nutrition, and improves fruit quality. Front. Plant Sci. 11, 764 (2020).
Zhang, J. et al. Variation and evolution of C:N ratio among different organs enable plants to adapt to N-limited environments. Glob. Change Biol. 26, 2534–2543 (2020).
An, J. et al. Dynamic regulation of anthocyanin biosynthesis at different light intensities by the BT2-TCP46-MYB1 module in apple. J. Exp. Bot. 71, 3094–3109 (2020).
Ji, X. et al. The BTB protein MdBT2 recruits auxin signaling components to regulate adventitious root formation in apple. Plant Physiol. 189, 1005–1020 (2022).
Gui, M. Y. et al. Studies of the relationship between rice stem composition and lodging resistance. J. Agric. Sci. 156, 387–395 (2018).
Tiwari, J. K. et al. Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci. Rep. 10, 1152 (2020).
Zhao, X. et al. The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic. Res. 6, 66 (2019).
Lian, X. et al. MdKCS2 increased plant drought resistance by regulating wax biosynthesis. Plant Cell Rep. 40, 2357–2368 (2021).
Yang, X. et al. The acyl desaturase CER17 is involved in producing wax unsaturated primary alcohols and cutin monomers. Plant Physiol. 173, 1109–1124 (2017).
Wang, Y. et al. Function and transcriptional regulation of CsKCS20 in the elongation of very-long-chain fatty acids and wax biosynthesis in Citrus sinensis flavedo. Hortic. Res 9, uhab027 (2022).
Sun, M. et al. The glucose sensor MdHXK1 phosphorylates a tonoplast Na+/H+ exchanger to improve salt tolerance. Plant Physiol. 176, 2977–2990 (2018).
Jiang, H. et al. The apple palmitoyltransferase MdPAT16 influences sugar content and salt tolerance via an MdCBL1–MdCIPK13–MdSUT2.2 pathway. Plant J. 106, 689–705 (2021).
Ma, Q. et al. A CIPK protein kinase targets sucrose transporter MdSUT2.2 at Ser254 for phosphorylation to enhance salt tolerance. Plant Cell Environ. 42, 918–930 (2019).
Han, P. et al. BTB-BACK domain E3 ligase MdPOB1 suppresses plant pathogen defense against Botryosphaeria dothidea by ubiquitinating and degrading MdPUB29 protein in apple. Plant Cell Physiol. 60, 2129–2140 (2019).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Acknowledgements
This work is supported by Natural Science Foundation of Shandong Province Outstanding Youth Fund Project (ZR2022JQ14), National Excellent Young Talents Support Program, National Natural Science Foundation of China (32072539), Taishan Scholar Youth Expert Project and Natural Science Foundation of Shandong Province Youth Fund Project (ZR2022QC112).
Author information
Authors and Affiliations
Contributions
Y.-Y.L., H.J. and C.-H.Q. planned and designed the research. H.J., C.-H.Q., H.-N.G., Z.-Q.F., Y.-T.W., X.-X.X., J.-Y.C., X.-F.W., Y.-H.L., W.-S.G., Y.-M.J. and C.-X.Y. performed experiments, conducted fieldwork, analysed data and so on. H.J., H.-N.G. and Y.-Y.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Cheng Chang, Yunjiang Cheng and Fabrizio Costa for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and Tables 1–5.
Source data
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, H., Qi, CH., Gao, HN. et al. MdBT2 regulates nitrogen-mediated cuticular wax biosynthesis via a MdMYB106-MdCER2L1 signalling pathway in apple. Nat. Plants 10, 131–144 (2024). https://doi.org/10.1038/s41477-023-01587-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01587-7