Abstract
Fruit lignification is due to lignin deposition in the cell wall during cell development. However, there are few studies on the regulation of cell wall lignification and lignin biosynthesis during fruit pigmentation. In this study, we investigated the regulation of cell wall lignification and lignin biosynthesis during pigmentation of winter jujube. The cellulose content decreased, while the lignin content increased in the winter jujube pericarp during pigmentation. Safranin O-fast green staining showed that the cellulose content was higher in the cell wall of winter jujube prior to pigmentation, whereas the lignin in the cell wall increased after pigmentation. The thickness of the epidermal cells decreased with pericarp pigmentation. A combined metabolomics and transcriptomics analysis showed that guaiacyl-syringyl (G-S) lignin was the main lignin type in the pericarp of winter jujube, and F5H (LOC107424406) and CCR (LOC107420974) were preliminarily identified as the key genes modulating lignin biosynthesis in winter jujube. Seventeen MYB and six NAC transcription factors (TFs) with potential regulation of lignin biosynthesis were screened out based on phylogenetic analysis. Three MYB and two NAC TFs were selected as candidate genes and further studied in detail. Arabidopsis ectopic expression and winter jujube pericarp injection of the candidate genes indicated that the MYB activator (LOC107425254) and the MYB repressor (LOC107415078) control lignin biosynthesis by regulating CCR and F5H, while the NAC (LOC107435239) TF promotes F5H expression and positively regulates lignin biosynthesis. These findings revealed the lignin biosynthetic pathway and associated genes during pigmentation of winter jujube pericarp and provide a basis for further research on lignin regulation.
Similar content being viewed by others
Introduction
Jujube (Ziziphus jujuba Mill.) is a medicinal and edible fruit that originates from China. Owing to various characteristics, such as tolerance of barren environments, salt-alkali conditions, drought, wind and sand, and early and quick harvesting, jujube trees are pioneers in promoting economic development and ecological construction in sand-alkali-arid regions1. Jujube fruit is nutritious and rich in a variety of functionally active components. Recent studies have found that ursolic acid and oleanolic acid in jujube fruit extracts inhibit breast cancer metastasis and cell apoptosis2. Moreover, polyphenols in the jujube pericarp were reported to effectively protect against myocardial ischemia caused by isoproterenol3.
Lignin is an important component of cell walls that maintains the rigidity of cell walls and provides mechanical support for the cells. Its hydrophobicity can prevent water loss. The secondary lignification of cell walls plays a key role in the growth and development of plants, as well as in the resistance to various biotic and abiotic stresses4. Lignin contains three monomers, namely, p-hydroxyphenyl lignin (H lignin), syringyl lignin (S lignin), and guaiacyl lignin (G lignin). Lignin biosynthesis is one of the most important flavonoid biosynthesis pathways, while the PAL, C4H, and 4CL genes serve as the common structural genes of the flavonoid and lignin biosynthesis pathways. Silencing of the PAL gene in tobacco resulted in decreased flavonoid and lignin contents and impaired plant growth and development5. Knockout of the C4H gene in Arabidopsis (Arabidopsis thaliana Heynh.) blocked the biosynthesis of flavonoid and lignin6. In addition, downregulating the expression of various genes, including cinnamoyl CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), and hydroxycinnamoyl-CoA (HCT), in Arabidopsis reduced the lignin content but increased the flavonoid content, and the plants were smaller with stunted development7,8.
Lignin biosynthesis is regulated by MYB TFs. Some MYB TFs can simultaneously regulate flavonoid and lignin synthesis in positive or negative directions. For instance, CmMYB8 in chrysanthemum (Dendranthema morifolium Tzvel.) simultaneously represses the accumulation of lignin and flavonoids9. However, most MYB TFs can only positively regulate flavonoid or lignin synthesis. For example, the overexpression of tomato SlMYB4 significantly reduced the expression levels of structural genes such as PAL, 4CL, C4H, and CCR and decreased lignin accumulation. In contrast, there was no significant change in the flavonoid content since SlMYB4 could not bind to the SlCHS1 promoter10. The overexpression of grape VvMYB5a and VvMYB5b in petunia upregulated the expression of the PAL, C4H, and 4CL genes, increased the content of anthocyanin, downregulated CCoAOMT1 expression, and reduced lignin content11. Furthermore, NAC is another specific TF in lignin biosynthesis. It has been reported that the fruit lignification of pear (Pyrus pyrifolia) is regulated by PpNAC187. Specifically, the overexpression of PpNAC187 upregulated the expression of the CCR and COMT genes and increased the content of lignin12. Another study showed that the overexpression of PdWND3A in poplar (Populus euphratica Oliv.) upregulated the expression of the F5H1 gene and promoted the biosynthesis of lignin13.
The pigmentation process of the jujube pericarp occurs with the ripening of the fruit and is accompanied by the deposition of insoluble fiber in the cell walls. However, studies on the regulation of cell wall lignification and lignin biosynthesis in jujube are lacking. Herein, we analyzed the changes in insoluble fiber and pericarp cells and the regulation of lignin biosynthesis during pigmentation of winter jujube (Z. jujuba Mill. cv. Dongzao) using biochemical, metabolomic, transcriptomic, and transgenic approaches.
Results
Changes in insoluble fiber contents and cell structures during the pigmentation process of winter jujube pericarp
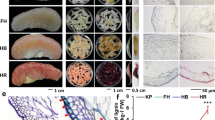
The pericarp color of winter jujube fruit at different periods of maturation varies from white (W) to semi-red (SR) to full-red (R) (Fig. 1A). As shown in Fig. 1B, cellulose was the main component in the pericarp of winter jujube, followed by hemicellulose and lignin. The cellulose content gradually decreased from the W stage to the R stage. In contrast, the hemicellulose and lignin contents of the winter jujube pericarp were the lowest in the W stage and increased by 52 and 91% in the R stage, respectively.
A Phenotypes of winter jujube in different coloring periods. B The contents of lignin, cellulose, and hemicellulose during the pigmentation of winter jujube. C Paraffin sections of pericarp using safranin O-fast green staining in different coloring periods. C1: pericarp of W, C2: white pericarp of SR, C3: red pericarp of SR, C4: pericarp of R. D Paraffin sections of pericarp in different coloring periods. D1: pericarp of W, D2: white pericarp of SR, D3: red pericarp of SR, D4: pericarp of R. E Layers and thickness of pericarp cells in different coloring periods. W: white stage; SR-W: white pericarp of the semi-red stage; SR-R: red pericarp of the semi-red stage; R: full-red stage
The epidermal cells of jujube were arranged closely and neatly at the W stage and SR-W stage, with oblate or oblong shapes (Fig. 1C). At the SR-R and R stages, the epidermal cells of winter jujube were significantly shriveled with increased transverse diameters and decreased longitudinal diameters with oblong shapes. Moreover, the intercellular space was increased, and the arrangement was looser than that in the prepigmentation stage (Fig. 1C). As shown in Fig. 1D, there was no color in the unstained jujube pericarp cells, while overall pericarp cells and cell walls displayed orange-yellow and orange-red colors, respectively, after pigmentation. Safranin O-fast green staining results (Fig. 1C) showed that the cell wall prior to pigmentation was blue-green in color with almost no color inside the cell. In contrast, the cells after pigmentation presented a red color, with an increased cell wall thickness. According to the principle of safranin O-fast green staining, safranin displayed the red color of the lignified cell wall, and fast green displayed the blue-green color of cellulose in the cell wall. Our results indicate that the cellulose content in the cell wall of winter jujube was high before pigmentation, while the lignin content was high when the cells became lignified after pigmentation.
As shown in Fig. 1E, the thickness of cuticle cells in the pericarp of winter jujube showed no significant change from the W to R stage, ranging from 9.3 to 10.2 μm. The thickness of the 5–6 layers of epidermal cells decreased with the pigmentation of the pericarp. Specifically, there was no significant change from the W stage (83.6 μm) to the SR-W stage (83.5 μm). However, there was a significant decrease to 59.7 μm at the SR-R stage and 61.2 μm at the R stage, demonstrating a decrease of 20 μm in thickness after pigmentation.
Observation of pigmentation on the pericarp of winter jujube
Figure 2 shows the anatomical structure of frozen sections of winter jujube pericarp at the R stage with six different thicknesses (5, 10, 15, 20, 25, and 30 μm). Consistent with the results from the paraffin sections, the color of the jujube pericarp adhered to the cuticle cells and 5–6 layers of epidermal cells. The epidermal cells of the jujube pericarp were oblong, with a large intercellular space and cellular pigmentation. As the section thickness increased, the color gradually darkened from yellow to orange-red. The sections in Fig. 2A (5 μm) and Fig. 2B (10 μm) were relatively thin and transparent, making it hard to capture the structural details. The sections in Fig. 2E (25 μm) and Fig. 2F (30 μm) were relatively thick, turbid, and inflexible, making it difficult to observe cellular structures. Ultimately, sections at thicknesses of 15 μm (Fig. 2C) and 20 μm (Fig. 2D) illustrated the pericarp cell structure and pigment deposition relatively clearly and vividly. We found through the observations that the epidermal cells of the red jujube pericarp were orange-red in color, with a large amount of pigments deposited on the cell walls.
A Frozen section of winter jujube pericarp at the R stage with 5 μm thicknesses. B Frozen section of winter jujube pericarp at the R stage with 10 μm thicknesses. C Frozen section of winter jujube pericarp at the R stage with 15 μm thicknesses. D Frozen section of winter jujube pericarp at the R stage with 20 μm thicknesses. E Frozen section of winter jujube pericarp at the R stage with 25 μm thicknesses. F Frozen section of winter jujube pericarp at the R stage with 30 μm thicknesses
Analysis of metabolites related to lignin biosynthesis in winter jujube pericarp during the pigmentation process
The accumulation of 13 major metabolites related to lignin biosynthesis in winter jujube during the pigmentation process is illustrated in Fig. 3A. Phenylalanine and p-coumaric acid levels were decreased as pigmentation increased. In contrast, p-coumaroyl, sinapyl aldehyde, and coniferyl aldehyde levels were higher in pigmented pericarps at the R and SR-R stages than in unpigmented pericarps at the W and SR-W stages. The contents of coniferyl alcohol and sinapyl alcohol first increased and then decreased, peaking at the SR-W stage. The p-coumaric alcohol content showed a declining trend, and its content was higher at the W stage than at the SR-W stage, with extremely low or no accumulation at the R and SR-R stages. These results indicate that G-S lignin rather than H-lignin is the main monomer of winter jujube pericarp.
A Analyses of metabolites involved in lignin biosynthesis during the pigmentation of winter jujube pericarp. Colors from blue to red indicate that the abundance of metabolites is low to high, and a redder color indicates that the abundance of differentially abundant metabolites is high. B Heatmap analyses of structural genes involved in lignin biosynthesis during the pigmentation of winter jujube pericarp. Colors from blue to red indicate that the levels of gene expression are low to high, and a redder color indicates that the levels of gene expression are high. W white stage, SR-W white pericarp of the semi-red stage, SR-R red pericarp of the semi-red stage, R: full-red stage
The gene expression levels of structural genes (PAL, C4H, 4CL, COMT, CCR, CAD, F5H, and POD) related to lignin biosynthesis in winter jujube were analyzed based on RNA-seq (Fig. 3B). Some genes were selected, and their expression levels were verified by qRT-PCR (Fig. S1). The results showed consistency between the transcriptome and qRT-PCR analysis. Specifically, the expression levels of PAL and C4H were downregulated during pericarp pigmentation. The expression levels of COMT genes (LOC107414981 and LOC107414981) were relatively high at the SR-R stage but low at the R stage. This pattern was different from the accumulating levels of sinapyl aldehyde, coniferyl aldehyde, and lignin. The expression levels of F5H (LOC107424406) and CCR (LOC107420974) were relatively high at the SR-R and R stages, consistent with the accumulation of sinapyl aldehyde, coniferyl aldehyde, and lignin. We deduced that F5H and CCR play key roles in G-S lignin biosynthesis. The expression of CAD (LOC107426959) was consistent with the accumulation of p-coumeric alcohol and may be the key gene for H-lignin biosynthesis.
Expression analysis of MYB TFs related to lignin biosynthesis
A phylogenetic tree was constructed with MYB TFs in jujube, Arabidopsis, and poplar to screen out the potential MYB TFs that regulate lignin biosynthesis in winter jujube. As shown in Fig. 4A, the results showed that the winter jujube MYB TFs LOC107417581, LOC107425254, and LOC107418291 were on the same evolutionary branch as poplar PtrMYB3 and PtrMYB2014 and Arabidopsis AtMYB4615, AtMYB8316, and AtMYB2617 (Group A3). LOC107403729 was on a branch with Arabidopsis AtMYB58 and AtMYB6318 (Group A2). LOC107434917 and LOC107414294 were on a branch with Arabidopsis AtMYB8519 (Group A1). LOC107430169 and LOC107430208 were on a branch with Arabidopsis AtMYB10320 (Group A4). LOC107424134, LOC107434709, LOC107434532, and LOC107406469 were on a branch with Arabidopsis AtMYB7519 (Group A7). LOC107432692 and LOC107415078 were on a branch with poplar PtrMYB621 (Group A5). LOC107421590, LOC107415776, and LOC107404478 were on a branch with poplar PtrMYB9322 (Group A6).
A Phylogenetic tree of 126 MYB TFs in Arabidopsis, 192 MYB TFs in poplar, and 99 MYB TFs in jujube. Accession numbers: AtMYB46 (AT5G12870), AtMYB83 (AT3G08500), AtMYB26 (AT3G13890), AtMYB58 (AT1G16490), AtMYB63 (AT1G79180), AtMYB75 (AT1G56650), AtMYB85 (AT4G22680), AtMYB103 (AT1G63910), PtrMYB3 (Potri.001G267300), PtrMYB20 (Potri.009G061500), PtrMYB6 (Potri.001G005100), and PtrMYB93 (Potri.004G138000.1). B Phylogenetic tree of 138 NAC TFs in Arabidopsis, 289 NAC TFs in poplar, and 101 NAC TFs in jujube. Accession numbers: AtNST1 (AT2G46770), AtNST2 (AT3G61910), AtNST3 (At1G32770), AtANAC012 (AT1G32770), AtANAC043 (AT2G46770), AtANAC066 (AT3G61910), AtANAC073 (AT4G28500), AtVNI1 (AT5G09330), AtVNI2 (AT5G13180), PtrWND1A (Potri.011G153300), PtrWND2A (Potri.014G104800), PtrWND1B (Potri.001G448400), and PtrWND2B (Potri.002G178700)
Seventeen jujube MYB TFs with potential regulation of lignin biosynthesis were screened out by phylogenetic analysis, among which nine TFs showed differential expression levels. The qRT-PCR results showed similar expression trends as the transcriptome analysis (Fig. 5A). In particular, only the expression of LOC107425254 was in accordance with changes in the lignin content, and the expression levels of LOC107415078 and LOC107415776 decreased with increasing lignin content.
A Heatmap displaying the expression levels (RNA-seq and qPCR) of candidate MYB genes involved in lignin biosynthesis during the pigmentation of winter jujube pericarp. B Heatmap displaying the expression levels (RNA-seq and qPCR) of candidate NAC genes involved in lignin biosynthesis during the pigmentation of winter jujube pericarp. Colors from blue to red indicate that the levels of gene expression are low to high, and a redder color indicates that the levels of gene expression are high. W white stage, SR-W white pericarp of the semi-red stage, SR-R red pericarp of the semi-red stage, R full-red stage
Expression analysis of NAC TFs related to lignin biosynthesis
To screen out the TFs associated with lignin biosynthesis in winter jujube from the NAC family, we used the amino acid sequences of 138 Arabidopsis NAC TFs23, 289 poplar NAC TFs (http://planttfdb.cbi.pku.edu.cn), and 101 jujube NAC TFs (http://planttfdb.cbi.pku.edu.cn/) to construct a phylogenetic tree. As shown in Fig. 4B, Arabidopsis AtNST124, AtNST2, AtNST3, ANAC012, ANAC043, and ANAC06625 and poplar PtrWND1A/2A and PtrWND1B/2B26 were located on the same branch, which was also shared by the jujube NAC TF LOC107423780 (Group B2/B3). Arabidopsis AtANAC07325 was on the same branch as jujube LOC107417198 and LOC107432797 (Group B4). Arabidopsis AtVNI227 was on the same branch as jujube LOC107435239 and LOC107417668 (Group B1). Arabidopsis AtVNI127 and jujube LOC107425600 were on the same branch (Group B5).
Six jujube NAC TFs associated with the regulation of lignin biosynthesis were screened out from the phylogenetic analysis. As shown in Fig. 5B, these six TFs were differentially expressed among samples and showed consistent expression levels between the transcriptome and qRT-PCR analysis, among which LOC107435239 and LOC107417668 exhibited relatively higher expression levels after jujube pigmentation, in line with the changes in lignin content.
Ectopic expression in Arabidopsis and pericarp injection in winter jujube
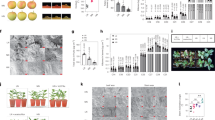
The above results identified three MYB (LOC107425254, LOC107415078, and LOC107415776) and two NAC (LOC107435239 and LOC107417668) TFs as candidates associated with lignin biosynthesis. These candidate genes were ectopically expressed in Arabidopsis and injected into the winter jujube pericarp. The cross sections of Arabidopsis stems stained with safranin O-fast green are shown in Fig. 6A. Overexpression of MYB (LOC107425254) or NAC (LOC107435239) in Arabidopsis led to deep staining of the stem (Fig. 6A), which was associated with increased lignin levels (Fig. 6D). However, only light staining of stems was observed with MYB (LOC107415078) overexpression (Fig. 6A), indicating a limited lignin level (Fig. 6D).
Significance tests are shown as i, ii, iii, and iv. Different letters above the columns denote significant differences (P < 0.01). A Cross sections of stems stained with safranin O-fast green in transgenic Arabidopsis. OE-pRI-101: overexpressing empty vectors. OE-MYB (LOC107425254)/(LOC107415078): overexpression of MYB (LOC107425254) or MYB (LOC107415078) in Arabidopsis, OE-NAC (LOC107435239): overexpression of NAC (LOC107435239) in Arabidopsis. B The winter jujube pericarp was injected with the mixed vectors and stored in a phytotron at 25 °C for 3 days. pIR; IL60-1 + IL60-2. pIR-MYB (LOC107425254): IL60-1 + MYB (LOC107425254)-IL60-2. pIR-MYB (LOC107415078): IL60-1 + MYB (LOC107415078)-IL60-2. pIR-NAC (LOC107415078): IL60-1 + NAC (LOC107435239)-IL60-2. C The injected winter jujube pericarp stained with safranin O-fast green. D Lignin contents of Arabidopsis stem. E Lignin contents in the winter jujube pericarp around the injection sites. F The expression of structural genes associated with lignin biosynthesis in the winter jujube pericarp around the injection sites
The injected winter jujube pericarps are shown in Fig. 6B, and the pericarps stained with safranin O-fast green are shown in Fig. 6C. The overexpression of MYB (LOC107425254) or NAC (LOC107435239) caused deep staining in the winter jujube pericarp around the injection sites, while overexpression of MYB (LOC107415078) resulted in only light staining. In addition, the lignin contents changed with the color of the staining (Fig. 6E). Further gene expression analysis showed that the overexpression of MYB (LOC107425254) in the winter jujube pericarp could significantly promote the expression of CCR (LOC107420974) and F5H (LOC107424406), while overexpression of NAC (LOC107435239) could only increase the expression of F5H (LOC107424406) (Fig. 6F). In contrast, the overexpression of MYB (LOC107415078) in the winter jujube pericarp reduced the expression of CCR (LOC107420974) and F5H (LOC107424406) (Fig. 6F).
Discussion
Changes in cell structure, cell wall lignification, and pigment deposition during the pigmentation process of winter jujube pericarp
Jujube pericarp pigmentation develops with the ripening process of fruits, which is very complex and involves many physiological processes, including cell senescence, pigment deposition, and metabolite accumulation. Cell senescence includes changes and modifications of cell wall components, as well as aging and death of organs28. Similarly, the cell structure of the jujube pericarp also changes with the pigmentation process, which has rarely been reported. Studies on the cell structure of jujube pericarp have mainly focused on the changes in cell structure during fruit cracking and fruit development29 rather than ripening and pigmentation. This study revealed that jujube pericarp cells are mainly composed of cuticle cells and epidermal cells (Fig. 1C, D). Moreover, the shape, size, and arrangement of epidermal cells varied with the ripening and pigmentation processes of the fruit. Specifically, the cell shape changed from oblate to oblong, and the arrangement changed from compact to relatively loose during the pigmentation process (Fig. 1C, D), which is consistent with the results of the previous research4. Moreover, it has been reported that the epidermal cell thickness and layer number of jujube fruits decrease with fruit ripening. In contrast, our study confirmed the decrease in epidermal cell thickness with fruit ripening, but the layer number remained unchanged (Fig. 1E). This discrepancy between the two studies may be associated with the difference in the jujube varieties used.
The lignification of the jujube pericarp is due to lignin deposition in the cell wall during cell development. Studies have shown organelle degradation, vacuole rupture, and cell content disappearance after cell wall lignification, leading to programmed cell death30. Zhao31 found that the contents of cellulose and hemicellulose in the pericarp decreased with fruit ripening. Li32 showed that the lignin biosynthetic pathway was activated at the transcriptional level during the coloration of green cotton fibers, indicating the function of lignin in pigmentation. In this study, our results showed a decrease in the cellulose content and an increase in the hemicellulose and lignin contents (Fig. 1B). The paraffin sections of jujube pericarp stained with safranin O-fast green revealed the lignification of pigmented epidermal cells reflected by the red color (Fig. 1C). In addition, frozen sections of jujube pericarp at the R stage showed that the epidermal cells were full of chromoplasts, with deposition of a large amount of pigments on the cell wall (Fig. 2). This phenomenon might be caused by the release of pigments (e.g., anthocyanins) from the cell vacuoles during cell wall lignification and cell apoptosis. Furthermore, the orange-red color of thickened cell walls might be associated with the high content of lignin or the polymerization of lignin with the spilled pigments into macromolecular phenols, which needs further verification.
Analysis of lignin biosynthesis and structural gene expression during the pigmentation process of winter jujube pericarp
It has been shown that the pericarp cells of Chinese jujube change with cell lignification, which is closely related to the accumulation of lignin33. Lignin consists of three monomers: H-lignin, G-lignin, and S-lignin. Generally, gymnosperms primarily have G-lignin, monocotyledons primarily have G-S-H lignin, and dicotyledons primarily have G-S lignin34. We studied the accumulation of 13 intermediate metabolites of the lignin biosynthesis pathway through metabolomics analysis, and the results showed that p-coumaric alcohol, the precursor of H-lignin, did not accumulate in the pericarp of winter jujube at the SR and R stages. Moreover, coniferyl alcohol, the precursor of G-lignin, and sinapyl alcohol, the precursor of S-lignin, were first increased and then decreased, with a peak at the SR stage (Fig. 3A), indicating the dominance of G-S lignin in the pericarp of winter jujube.
There are eight main structural genes involved in the lignin biosynthesis pathway35, among which PAL, C4H, and 4CL are the pro-structural genes for lignin biosynthesis. COMT and F5H are the key genes for G-S lignin biosynthesis. In alfalfa, downregulation of COMT expression reduced the content of G-lignin, while S-lignin remained unchanged36. Takeda et al.37 overexpressed F5H in rice to increase the S-lignin content. Moreover, overexpression of BpCCR1 in birch increased lignin content by 14.6%, while knockdown of this gene lowered the lignin content by 6.3%38. In our study, the expression levels of COMT genes (LOC107414981 and LOC107414981) were relatively high at the SR_R stage but low at the R stage, which was not consistent with the accumulation of coniferyl aldehyde, sinapyl aldehyde, and lignin (Fig. 3 and Fig. S1). The expression levels of F5H (LOC107424406) and CCR (LOC107420974) were relatively high at the SR_R and R stages, which was consistent with the accumulation of coniferyl aldehyde, sinapyl aldehyde, and lignin (Fig. 3 and Fig. S1). The results indicated that F5H (LOC107424406) and CCR (LOC107420974) might be the key genes for lignin biosynthesis in the pericarp of winter jujube. Notably, we also found that the expression of CAD (LOC107426959) was consistent with the accumulation of p-coumaric alcohol (Fig. 3), which was in line with a previous report that CAD was one of the specific enzymes in the lignin biosynthesis pathway, and the loss of CAD activity impacted lignin composition more than lignin content39.
Regulation of TFs related to lignin during the pigmentation process of winter jujube pericarp
MYB TFs are important regulators of lignin metabolism. Some MYB TFs participate in the regulation of the formation and lignification of secondary cell walls. For example, overexpression of AtMYB46 and AtMYB83 activated related structural genes in the lignin biosynthesis pathway, resulting in lignin accumulation and secondary cell wall thickening in Arabidopsis15,16. In this study, we revealed that the MYB TF LOC107425254, which was on the same branch as AtMYB46 and AtMYB83 (Fig. 4A), was upregulated with increasing lignin content in the winter jujube pericarp (Fig. 5A), indicating positive regulation of the lignification of winter jujube pericarp cells. Increased lignin content was detected in Arabidopsis or winter jujube pericarp overexpressing MYB (LOC107425254) (Fig. 6A–E). Further analysis showed that it could significantly promote the expression of CCR (LOC107420974) and F5H (LOC107424406) (Fig. 6F), suggesting that MYB (LOC107425254) positively controls lignin content by regulating CCR (LOC107420974) and F5H (LOC107424406).
PtrMYB6 negatively regulated lignin biosynthesis in poplars. Specifically, reduced lignin content and the number of cell layers, as well as thinner cell walls, were observed in the transgenic plants overexpressing PtrMYB621. Another study also showed that poplars overexpressing PtrMYB93 had lower lignin contents. Alternatively, when PtrMYB93 was knocked out, the expression levels of key structural genes in the lignin biosynthesis pathway were significantly upregulated, and the lignin content was increased22. In this study, we found that the levels of the MYB TFs LOC107415776 and LOC107415078 were downregulated with increasing lignin content in the winter jujube pericarp (Fig. 5A). Phylogenetic analysis showed that LOC107415776 and PtrMYB6 were on the same branch (Fig. 4A), while LOC107415078 and PtrMYB93 were on the same branch (Fig. 5A), indicating that they negatively regulate lignin biosynthesis. However, in this study, only MYB (LOC107415078) was predicted to negatively regulate lignin biosynthesis by reducing the expression of CCR (LOC107420974) and F5H (LOC107424406) (Fig. 6).
NAC TFs, such as AtNST1/2/324,25 and AtVNI1/227 of Arabidopsis and PtrWND1A/2A and PtrWND1B/2B of poplar26, also play an important role in lignin biosynthesis. In this study, two NAC TFs, LOC107435239, and LOC107417668 were selected for analysis, and their expression levels were upregulated with increasing lignin content (Fig. 5B). Phylogenetic analysis indicated that they were on the same branch as AtVNI2 (Fig. 4B). AtVNI2 can inhibit the activation of the AtVDN7 TF, which specifically regulates the development of secondary cell wall27. Therefore, we speculated that LOC107435239 and LOC107417668 might be involved in the metabolic process of secondary cell wall development. Only the overexpression of NAC (LOC107435239) increased lignin content in Arabidopsis mutants (Fig. 6A, D). Further analysis also showed that the overexpression of NAC (LOC107435239) positively regulated lignin biosynthesis in the winter jujube pericarp around the injection sites (Fig. 6B–E) and significantly promoted the expression of F5H (LOC107424406) (Fig. 6F).
Conclusion
A regulatory model of lignin biosynthesis in winter jujube pericarp during pigmentation is proposed based on the results from this study (Fig. 7). With the increase in lignin content and the lignification of cell walls during pigmentation of winter jujube pericarp, the thickness of the epidermal cells decreased. The lignin of the winter jujube pericarp was mainly G-S lignin, and F5H (LOC107424406) and CCR (LOC107420974) were preliminarily identified as the key genes for G-S lignin biosynthesis. This study also identified an MYB activator (LOC107425254) and an MYB repressor (LOC107415078) associated with lignin biosynthesis by regulating CCR (LOC107420974) and F5H (LOC107424406), while the NAC (LOC107435239) TF significantly promoted the expression of F5H (LOC107424406) and positively regulated lignin biosynthesis. In conclusion, these results revealed the metabolic pathway and key genes controlling lignin biosynthesis during pigmentation of winter jujube pericarp and provide a basis for further research on lignin regulation.
Materials and methods
Plant materials
Winter jujube fruits were collected from the germplasm repository of Shandong Institute of Pomology in 2018. The repository was located in the Taishan District of Tai’an City, Shandong Province, China. The growth conditions were a semihumid continental monsoon climate zone, a plain region with clay loam soils, with an average annual temperature of 13 °C, average annual precipitation of 697 mm, and a frost-free period of 195 days. Three 13-year-old healthy jujube plants with similar growth statuses and under unified management were selected for the study. Briefly, ten jujube fruits with homogeneous size, shape, and color were randomly collected from different locations of the sample trees at the white stage (W), semi-red stage (SR), and full-red stage (R). Fruit samples with infection of diseases and pests or mechanical damage were strictly excluded from the study. Different samples were mixed as a biological replicate and used in triplicate for each experiment. After collection, samples were immediately brought back to the laboratory, washed, peeled, frozen in liquid nitrogen, and stored at −80 °C for subsequent uses.
Determination of insoluble fiber
The lignin content was determined according to the method of Shilton40 with slight modification. Briefly, 0.5 g of sample was homogenized with 95% ethanol and centrifuged at 4000 rpm for 10 min. The pellet was washed three times with 95% ethanol and then rinsed three times with ethanol:n-hexane (1:1) solution, and the precipitate was collected and dried. The dry mass was placed in the test tube, dissolved in 0.5 mL of 25% bromoacetyl, and incubated at 70 °C in a water bath for 30 min, followed by the addition of 0.9 mL of 2 mol ∙ L−1 NaOH to terminate the reaction. Subsequently, 5 mL of glacial acetic acid and 0.1 mL of 7.5 mol ∙ L−1 hydroxylamine hydrochloride were added. The mixture was centrifuged for 5 min, 0.1 mL of supernatant was taken and diluted by adding 3.0 mL of glacial acetic acid, and the absorbance value at 280 nm was determined.
The cellulose content was measured by the Cellulase Assay Kit according to the manufacturer’s instructions. Specifically, 0.3 g of sample was first incubated in 80% ethanol for 20 min at 95 °C. After cooling, the sample was centrifuged at 4000 rpm for 10 min. The pellets were vortexed with 1.5 ml of 80% ethanol for 2 min, followed by 2 min of centrifugation at 4000 rpm. After removal of ethanol, the sample was washed with acetone, followed by incubation with 1 mL of reagent I for 15 h. Samples were centrifuged, and the pellets were dried in an oven at 105 °C to constant weights. Five milligrams of dried sample was homogenized using 0.5 mL of deionized water and incubated on ice. Slowly, 0.75 mL of concentrated sulfuric acid was added, and the samples were incubated for another 30 min. The supernatant after centrifugation at 4000 rpm for 10 min was taken and diluted 20 times using deionized water for subsequent measurement. For the preparation of the working solution, 4 mL of reagent III was added into reagent II. Seventy microliters of working solution and 630 μL of concentrated sulfuric acid were added to 300 μL of the prepared sample solution and deionized water, respectively, and incubated at 95 °C for 10 min, and the absorbance at 620 nm was recorded after cooling.
The hemicellulose content was determined by using a commercial hemicellulose detection kit. According to the manufacturer’s instructions, 2 mL of reagent I was added to 0.1 g of the sample, incubated at 90 °C for 10 min, and centrifuged at 5000 rpm for 10 min. The pellets were washed three times with 1 mL of deionized water and kept in an oven at 105 °C until they reached a constant weight, followed by the addition of 1 mL of reagent II. The mixture was incubated at 90 °C for 1 h, followed by the addition of 0.1 mL of reagent III and 1 mL of reagent IV and mixing until the color was reddish. The solution was centrifuged at 5000 rpm for 10 min, and 0.2 mL of the supernatant was thoroughly mixed with 0.15 mL of reagent V and 0.65 mL of deionized water. After 5 min of incubation in a water bath at 90 °C, absorbance at 540 nm was recorded.
Preparation and observation of paraffin sections
Pericarp samples with a thickness less than 0.2 cm were sliced from the winter jujube fruits from the W stage, SR-W stage, SR-R stage, and R stage cut into 0.3 cm × 0.3 cm pieces, fixed with FAA fixing solution, and incubated in 70% ethanol solution for 12 h. Furthermore, the pericarp samples were dehydrated consecutively in 80% ethanol for 60 min, 90% ethanol for 30 min, and 100% ethanol for 15 min. The dehydrated samples were further incubated sequentially in anhydrous ethanol/xylene solution for 15 min, in xylene solution for 3 min, and in paraffin/xylene for 30 min, followed by paraffin embedding for 80 min. The embedded samples were sliced into sections with a thickness of 5 μm. The paraffin slices were picked out with toothpicks and placed on slides, developed with warm water at 35 °C, placed on a slide coated with a thin layer of protein glycerin, and dried in an oven at 38 °C.
The samples at the same stage were then treated with safranin O-fast green staining as follows: the sections were placed in xylene I for 20 min, xylene II for 20 min, anhydrous ethanol I for 5 min, anhydrous ethanol II for 5 min, and 75% ethanol for 5 min, and then rinsed with ultrapure water. The treated samples were stained in 0.1% safranin solution for 2 h, rinsed with ultrapure water, and placed in 50, 70, and 80% ethanol for 5 s. The samples were then stained with 0.15% solid green dye for 60 s, rinsed with ultrapure water and glacial acetic acid solution, dehydrated with 95% ethanol for 10 s, dehydrated with anhydrous ethanol for 10 s, processed in xylene for 5 min, and sealed with neutral glue. Nonstained and stained slices were observed under an optical microscope (Olympus BX53) and photographed. Changes in the epidermal cell layer and cell structure of the jujube pericarp were observed. The thicknesses of cuticle and epidermal cells were measured with CaseViewer software (The Digital Pathology Company).
Preparation and observation of frozen sections
Fresh winter jujube fruits at the R stage were cut into 0.5 cm × 0.5 cm × 0.2 cm pieces, placed in the sample disc, and surrounded by OCT embedding drops. The sample disc was slowly placed into the liquid nitrogen tank. When the disc bottom reached the liquid nitrogen, the disc was held for 10–20 s before being immersed in liquid nitrogen. The frozen samples were sliced into sections with thicknesses of 5, 10, 15, 20, 25, and 30 μm. The sections were observed and photographed under an optical microscope (Olympus BX53). The number of epidermal cell layers, the change in cell structures, and pigment deposition were recorded.
Ultraperformance liquid chromatography-mass spectrometry (UPLC-MS) analysis
Approximately 0.1 g of winter jujube pericarp sample was ground into powder in liquid nitrogen, and 500 μL of 80% methanol solution containing 0.1% formic acid was added; then, the sample was vortexed and placed in an ice bath for 5 min. After centrifugation at 15,000 rpm for 10 min at 4 °C, the sample was diluted with a 53% methanol solution and centrifuged at 15,000rpm for 20 min at 4 °C. The supernatant was used for UPLC-MS analysis. Aliquots from each test sample were mixed as quality control (QC) samples.
The UPLC assay was performed on a Shim-pack Shimadzu CBM30A equipped with a Waters Acquity UPLC HSS T3 C18 column of 1.8 µm (2.1 mm × 100 mm). The experimental conditions were as follows. flow rate: 0.4 ml min−1; column temperature: 40 °C; injection volume: 2 μL; mobile phase A: 0.04% acetic acid aqueous solution, mobile phase B: 0.04% acetonitrile solution. Sample elution was established by mixing mobile phase A/mobile phase B (V/V) in a linear gradient: 0 min was 95:5, 11.0 min was 5:95, 12.0 min was 5:95, 12.1 min was 95:5, and 15.0 min was 95:5.
Mass spectrum analysis was carried out by a triple four-pole tandem mass spectrometer (Applied Biosystems 6500 QTRAP) with the following parameters: temperature 500 °C, voltage 5500 V, curtain gas (CUR) 25 psi, and collision-activated dissociation (CAD) as high.
Qualitative and quantitative analysis of metabolites and data analysis
Based on the Metware Database (MWDB) and other public metabolite information databases, metabolite characterization was carried out according to the secondary spectral information. During the analysis, isotope signals, repeated K+, Na+, and NH4+ ion signals, and repetitive signals of fragment ions contained in other larger molecules were removed. Metabolites were quantitatively analyzed by multiple reaction monitoring (MRM) using Triple Quad mass spectrometry.
The metabolite data were analyzed using Analyst 1.6.1 software (AB Sciex, Ontario, Canada). Principal component analysis (PCA) was used to analyze the differences in population metabolism among samples and the degree of variation among samples within a group. The metabolites were analyzed by partial least square discriminant analysis (OPLS-DA), and the metabolites with variable importance in projection (VIP) ≥1 and P value ≤0.5 were identified as differentially abundant metabolites. Hierarchical clustering analysis (HCA) and metabolite correlation analysis were used to reveal the relationship between metabolites and samples.
Transcriptome analysis
Total RNA was extracted from the pericarp of winter jujube using an RNAprep Pure Polysaccharide polyphenol total plant RNA extraction kit (TransGen Biotech, Beijing, China). The qualified total RNA was used for subsequent library construction and sequencing by the Illumina NovaSeq 6000 platform.
The reference genome sequence and functional gene annotation files were retrieved from https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/826/755/GCF_000826755.1_ZizJuj_1.1. Hisat2 V2.0.5 was used to compare clean paired-end reads with the reference genome. Reads of each gene in each sample were calculated using featureCounts V1.5.0-p3, and the value of fragments per kilobase of exon per million mapped reads (FPKM) of each gene was calculated based on the length of the gene.
DESeq2 R software (version 1.16.1) was used to compare expression differences between paired groups. Genes with P < 0.05 and | log2(fold change) | >0 after correction were considered significantly differentially expressed genes for subsequent analysis.
Gene Ontology (GO) enrichment and KEGG enrichment analysis of differentially expressed genes were achieved by clusterProfiler R software, in which the gene length bias was corrected. GO terms and KEGG terms with adjusted P < 0.05 were used as significantly enriched functional terms for subsequent analysis.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The selected genes and TFs were verified by qRT-PCR. Specific primers were designed using Primer 6.0 according to the whole genome sequences of winter jujube. The TransStart® Top Green qPCR SuperMix kit was used for detection. Each sample was analyzed in triplicate, with ZjActin used as the internal reference gene. The Ct values were read under default conditions, and the 2−ΔΔCT method was used for data analysis41.
Ectopic expression in Arabidopsis and pericarp injection of winter jujube
Ectopic expression was carried out as described by Wang et al.42, with slight alterations. The recombinant plasmids containing candidate genes were transformed into Agrobacterium tumefaciens GV3101, which was utilized to infect Arabidopsis. The T1 transgenic Arabidopsis plants were then selected by plating onto MS medium containing hygromycin. The hygromycin-resistant seedlings were moved to soil and grown in a growth chamber (Ningbo-Jiangnan, http://www.nbjnyq.com/). The T2 seeds were then collected and grown as described above, and the lignin contents of their stems were determined.
Pericarp injection assays were carried out as described previously43. The overexpression of viral vectors MYB (LOC107425254)-IL60–2, MYB (LOC107415078)-IL60-2, and NAC (LOC107435239)-IL60-2 was generated by inserting the corresponding CDSs into the IL60-2 vector. The IL60-1 vector was used as an auxiliary plasmid. The mixed vectors (IL60-1: IL60-2 = 1: 1; 500 μL: 100 μM acetosyringone, 10 mM 2-morpholinoethanesulfonic acid) were injected into the fruit pericarp using a 1 mL medical syringe. Then, the fruits were stored in a phytotron at 25 °C for 3 days.
Statistical analysis
All results shown represent the mean of three independent assays. Significant differences between groups were determined by Duncan’s new multiple range test, with significance tests denoted as i, ii, iii, and iv. Different lowercase letters in figures and tables denote significant differences (P < 0.05).
Data availability
Supporting data not included in the manuscript are stored in (https://pan.baidu.com/share/init?surl=G_N60yxSOw-b4zKyHsNMgg, password: 2021).
References
Liu, M. J., Wang M. Germplasm Resources of Winter jujube [M] (China Forestry Publishing House, 2009).
Tahergorabi, Z. et al. “Ziziphus jujuba”: a red fruit with promising anticancer activities. Pharmacogn. Rev. 9, 99–106 (2015).
Cheng, D., Cao, J. & Jiang, W. The protective effects of polyphenols from jujube peel (Ziziphus Jujuba Mill) on isoproterenol-induced myocardial ischemia and aluminum-induced oxidative damage in rats. Food Chem. Toxicol. 50, 1302–1308 (2012).
Boerjan, W., Ralph, J. & Baucher, M. Lignin biosynthesis. Annu Rev. Plant Biol. 54, 519–546 (2003).
Korth, K. L. et al. Changes in phenylpropanoid metabolites associated with somatic reversion of homology-dependent silencing of phenylalanine ammonia-lyase in tobacco. Physiol. Plant. 111, 137–143 (2008).
Schilmiller, A. et al. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60, 771–782 (2009).
Besseau, S. et al. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19, 148–162 (2007).
Thbenin, J. et al. The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 4, 70–82 (2011).
Zhu, L. et al. CmMYB8 encodes an R2R3 MYB transcription factor which represses lignin and flavonoid synthesis in chrysanthemum. Plant Physiol. Biochem. 149, 217–224 (2020).
Yang, C. Study on the Mechanism of Regulating Tomato Lignin Synthesis [D] (Huazhong Agricultural Univ., 2019).
Cavallini, E. et al. Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant[J]. Plant Cell Physiol. 55, 517–534 (2014).
Li, M. et al. PpNAC187 enhances lignin synthesis in ‘Whangkeumbae’ Pear (Pyrus pyrifolia) ‘hard-end’ fruit. Molecules 24, 4338 (2019).
Yang, Y. et al. PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in Populus. BMC Plant Biol. 19, 486 (2019).
Mccarthy, R. L. et al. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090 (2010).
Zhong, R., Richardson, E. A. & Ye, Z. H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792 (2007).
Mccarthy, R. L., Zhong, R. & Ye, Z. H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964 (2009).
Yang, C. et al. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548 (2007).
Zhou, J. et al. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266 (2009).
Zhong, R. et al. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782 (2008).
Ohman, D. et al. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 73, 63–76 (2013).
Wang, L. et al. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 99, 733–751 (2019).
Dou, L W. The Negative Regulational Molecular Mechanism of the Poplar Transcription Factor MYB93 Involved in Flavonoids and Lignin Biosynthesis [D] (Southwest Univ., 2019).
Ooka, H. et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247 (2003).
Mitsuda, N. et al. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006 (2005).
Ko, J. H. et al. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 50, 1035–1048 (2007).
Ohtani, M. et al. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 67, 499–512 (2011).
Yamaguchi, M. et al. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263 (2010).
Piccolo, E. L. et al. Girled-induced anthocyanin accumulation in red-leafed Prunus cerasifera: effect on photosynthesis, photoprotection and sugar metabolism. Plant Sci. 294, 110456 (2020).
Wu, C. et al. Effect of ripening stage on physicochemical properties and antioxidant profiles of a promising table fruit “pear-jujube” (Zizyphus jujuba Mill.). Sci. Hortic. 148, 177–184 (2012).
Huang, J. et al. Jujube cultivar vitamin C profile and nutrient dynamics during maturation. Am. Soc. Hortic. Sci. 52, 859–867 (2017).
Zhao, L. Q. Effect of Organizational Structure and Material Changes on Jujube Fruit Cracking in Growth and Development Process [D] (Shanxi Agricultural Univ., 2013).
Li, Z. H. et al. Phenylpropanoid metabolism and pigmentation show divergent patterns between brown color and green color cottons as revealed by metabolic and gene expression analyses. J. Cotton Res. 3, 27 (2020).
Mahabir, S. & Narender, S. Impact of lignification on secondary cell wall development: a review. Asian J. Plant Sci. Res 9, 30–37 (2019).
Rogers, L. A. & Campbell, M. M. The genetic control of lignin deposition during plant growth and development. N. Phytol. 164, 17–30 (2004).
Weng, J. K. & Chapple, C. The origin and evolution of lignin biosynthesis. N. Phytol. 187, 273–285 (2010).
Guo, D. et al. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-Methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S Lignin. Plant Cell 13, 73–88 (2001).
Takeda, Y. et al. Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 246, 337–349 (2017).
Zhang, W. et al. Functional characterization of CCR in birch (Betula platyphylla × Betula pendula) through overexpression and suppression analysis. Physiol. Plant 154, 283–296 (2015).
Hplpin, R. W. et al. Manipulation of lignin quality by down regulation of cinnamyl alcohol dehydrogenase. Plant J. 6, 121–129 (1994).
Shilton, B. The effect of plant hormones on phenolic production in Kauri trees. MSc thesis, Auckland University of Technology (2017).
Kenneth, J. L. & Thomas, D. S. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 25, 402–408 (2001).
Wang, N. et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292 (2017).
An, J. P. et al. Dynamic regulation of anthocyanin biosynthesis at different light intensities by the BT2-TCP46-MYB1 module in apple. J. Exp. Bot. 71, 3094–3109 (2020).
Acknowledgements
The authors would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript. This work was supported by the National Key R&D Program of China (2019YFD1001605), the Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2017D01 and CXGC2018E22), Shandong Academy of Agricultural Sciences introduction and training of high-level talents (1-18-078), the Science and Technology Research Project of University in Hebei Province (QN2020205), the Shandong Key Research and Development Program (2016GNC113015 and 2019GNC106061), the Shandong Agricultural Applied Technology Innovation Project (2017), and the Shandong Provincial Natural Science Fund (ZR2020QC141).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Q., Wang, L., Wang, Z. et al. The regulation of cell wall lignification and lignin biosynthesis during pigmentation of winter jujube. Hortic Res 8, 238 (2021). https://doi.org/10.1038/s41438-021-00670-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00670-4
This article is cited by
-
Gene Expression Pattern, Lignin Deposition and Root Cell Wall Modification of Developing Mangrove Propagules Under Salinity Stress
Journal of Plant Growth Regulation (2023)
-
Evolution and Analysis of Caffeic Acid Transferase (COMT) in Seed Plants
Biochemical Genetics (2023)
-
Transcriptomic and metabolomic profiling of flavonoid biosynthesis provides novel insights into petals coloration in Asian cotton (Gossypium arboreum L.)
BMC Plant Biology (2022)