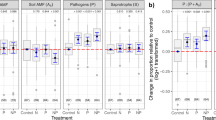
Abstract
Plant–soil feedbacks (PSFs), soil-mediated plant effects on conspecific or heterospecific successors, are a major driver of vegetation development. It has been proposed that specialist plant antagonists drive differences in PSF responses between conspecific and heterospecific plants, whereas contributions of generalist plant antagonists to PSFs remain understudied. Here we examined PSFs among nine annual and nine perennial grassland species to test whether poorly defended annuals accumulate generalist-dominated plant antagonist communities, causing equally negative PSFs on conspecific and heterospecific annuals, whereas well-defended perennial species accumulate specialist-dominated antagonist communities, predominantly causing negative conspecific PSFs. Annuals exhibited more negative PSFs than perennials, corresponding to differences in root–tissue investments, but this was independent of conditioning plant group. Overall, conspecific and heterospecific PSFs did not differ. Instead, conspecific and heterospecific PSF responses in individual species’ soils were correlated. Soil fungal communities were generalist dominated but could not robustly explain PSF variation. Our study nevertheless suggests an important role for host generalists as drivers of PSFs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Fungal ITS2 sequence data are uploaded to NCBI under project number PRJNA952944. Plant biomass data of the main experiment as well as root trait data are available on figshare under: https://doi.org/10.6084/m9.figshare.22740974 ref. 69. The FUNGUILD database46 (http://www.funguild.org) was used for fungal ASV annotation to ecological guilds. Source data are provided with this paper.
References
Lekberg, Y. et al. Relative importance of competition and plant-soil feedback, their synergy, context dependency and implications for coexistence. Ecol. Lett. 21, 1268–1281 (2018).
De Deyn, G. B. et al. Soil invertebrate fauna enhances grassland succession and diversity. Nature 422, 711–713 (2003).
Liu, S. et al. Phylotype diversity within soil fungal functional groups drives ecosystem stability. Nat. Ecol. Evol. 6, 900–909 (2022).
Wilschut, R. A. et al. Root traits and belowground herbivores relate to plant–soil feedback variation among congeners. Nat. Commun. 10, 1564 (2019).
Philippot, L., Raaijmakers, J. M., Lemanceau, P. & van der Putten, W. H. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799 (2013).
Bergmann, J. et al. The fungal collaboration gradient dominates the root economics space in plants. Sci. Adv. 6, eaba3756 (2020).
Bezemer, T. M. et al. Divergent composition but similar function of soil food webs of individual plants: plant species and community effects. Ecology 91, 3027–3036 (2010).
van der Putten, W. H. et al. Plant–soil feedbacks: the past, the present and future challenges. J. Ecol. 101, 265–276 (2013).
Mangan, S. A. et al. Negative plant–soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–755 (2010).
Klironomos, J. N. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70 (2002).
Kempel, A., Rindisbacher, A., Fischer, M. & Allan, E. Plant soil feedback strength in relation to large-scale plant rarity and phylogenetic relatedness. Ecology 99, 597–606 (2018).
Bennett, J. A. et al. Plant–soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355, 181–184 (2017).
Kardol, P., Bezemer, T. M. & van der Putten, W. H. Temporal variation in plant–soil feedback controls succession. Ecol. Lett. 9, 1080–1088 (2006).
van der Putten, W. H., Van Dijk, C. & Peters, B. A. M. Plant-specific soil-borne diseases contribute to succession in foredune vegetation. Nature 362, 53–56 (1993).
Thakur, M. P. et al. Plant–soil feedbacks and temporal dynamics of plant diversity–productivity relationships. Trends Ecol. Evol. 36, 651–661 (2021).
van de Voorde, T. F. J., van der Putten, W. H. & Martijn Bezemer, T. Intra- and interspecific plant–soil interactions, soil legacies and priority effects during old-field succession. J. Ecol. 99, 945–953 (2011).
Kardol, P., Cornips, N. J., van Kempen, M. M. L., Bakx-Schotman, J. M. T. & van der Putten, W. H. Microbe-mediated plant-soil feedback causes historical contingency effects in plant community assembly. Ecol. Monogr. 77, 147–162 (2007).
Callaway, R. M., Montesinos, D., Williams, K. & Maron, J. L. Native congeners provide biotic resistance to invasive Potentilla through soil biota. Ecology 94, 1223–1229 (2013).
Kulmatiski, A., Beard, K. H., Stevens, J. R. & Cobbold, S. M. Plant–soil feedbacks: a meta-analytical review. Ecol. Lett. 11, 980–992 (2008).
Semchenko, M. et al. Deciphering the role of specialist and generalist plant–microbial interactions as drivers of plant-soil feedback. New Phytol. 234, 1929–1944 (2022).
Spear, E. R. & Broders, K. D. Host-generalist fungal pathogens of seedlings may maintain forest diversity via host-specific impacts and differential susceptibility among tree species. New Phytol. 231, 460–474 (2021).
Van der Putten, W. H. Plant defense belowground and spatiotemporal processes in natural vegetation. Ecology 84, 2269–2280 (2003).
Mommer, L. et al. Lost in diversity: the interactions between soil-borne fungi, biodiversity and plant productivity. New Phytol. 218, 542–553 (2018).
Ruijven, J., Ampt, E., Francioli, D., Mommer, L. & Fridley, J. Do soil‐borne fungal pathogens mediate plant diversity–productivity relationships? Evidence and future opportunities. J. Ecol. 108, 1810–1821 (2020).
Wilschut, R. A. & Geisen, S. Nematodes as drivers of plant performance in natural systems. Trends Plant Sci. 26, 237–247 (2021).
Cortois, R. et al. Plant–soil feedbacks: role of plant functional group and plant traits. J. Ecol. 104, 1608–1617 (2016).
Semchenko, M. et al. Fungal diversity regulates plant-soil feedbacks in temperate grassland. Sci. Adv. 4, eaau4578 (2018).
Lemmermeyer, S., Lorcher, L., van Kleunen, M. & Dawson, W. Testing the plant growth–defense hypothesis belowground: do faster-growing herbaceous plant species suffer more negative effects from soil biota than slower-growing ones? Am. Nat. 186, 264–271 (2015).
Xi, N. et al. Relationships between plant–soil feedbacks and functional traits. J. Ecol. 109, 3411–3423 (2021).
Dowarah, B., Gill, S. S. & Agarwala, N. Arbuscular mycorrhizal fungi in conferring tolerance to biotic stresses in plants. J. Plant Growth Regul. 41, 1429–1444 (2021).
Jarosz, A. M. & Davelos, A. L. Effects of disease in wild plant populations and the evolution of pathogen aggressiveness. New Phytol. 129, 371–387 (2006).
Spitzer, C. M. et al. Root traits and soil micro‐organisms as drivers of plant–soil feedbacks within the sub‐arctic tundra meadow. J. Ecol. 110, 466–478 (2021).
Grime, J. P. Plant Strategies, Vegetation Processes, and Ecosystem Properties (Wiley, 2006).
Bennett, J. A. & Klironomos, J. Mechanisms of plant–soil feedback: interactions among biotic and abiotic drivers. New Phytol. 222, 91–96 (2019).
De Long, J. R. et al. Contrasting responses of soil microbial and nematode communities to warming and plant functional group removal across a post-fire boreal forest successional gradient. Ecosystems 19, 339–355 (2015).
Olff, H., Hoorens, B., de Goede, R. G. M., van der Putten, W. H. & Gleichman, J. M. Small-scale shifting mosaics of two dominant grassland species: the possible role of soil-borne pathogens. Oecologia 125, 45–54 (2000).
Vincenot, C. E., Cartenì, F., Bonanomi, G., Mazzoleni, S. & Giannino, F. Plant–soil negative feedback explains vegetation dynamics and patterns at multiple scales. Oikos 126, 1319–1328 (2017).
in 't Zandt, D. et al. Species abundance fluctuations over 31 years are associated with plant–soil feedback in a species‐rich mountain meadow. J. Ecol. 109, 1511–1523 (2020).
Mordecai, E. A. Pathogen impacts on plant diversity in variable environments. Oikos 124, 414–420 (2015).
Lepinay, C., Vondrakova, Z., Dostalek, T. & Munzbergova, Z. Duration of the conditioning phase affects the results of plant–soil feedback experiments via soil chemical properties. Oecologia 186, 459–470 (2018).
Maron, J. L., Marler, M., Klironomos, J. N. & Cleveland, C. C. Soil fungal pathogens and the relationship between plant diversity and productivity. Ecol. Lett. 14, 36–41 (2011).
Veen, G. F. et al. The role of plant litter in driving plant–soil feedbacks. Front. Environ. Sci. 7, 168 (2019).
Lekberg, Y. et al. More bang for the buck? Can arbuscular mycorrhizal fungal communities be characterized adequately alongside other fungi using general fungal primers? New Phytol. 220, 971–976 (2018).
Benitez, M. S., Hersh, M. H., Vilgalys, R. & Clark, J. S. Pathogen regulation of plant diversity via effective specialization. Trends Ecol. Evol. 28, 705–711 (2013).
Nilsson, R. H. et al. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17, 95–109 (2019).
Nguyen, N. H. et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016).
Tedersoo, L. & Anslan, S. Towards PacBio-based pan-eukaryote metabarcoding using full-length ITS sequences. Environ. Microbiol. Rep. 11, 659–668 (2019).
Eck, J. L., Stump, S. M., Delavaux, C. S., Mangan, S. A. & Comita, L. S. Evidence of within-species specialization by soil microbes and the implications for plant community diversity. Proc. Natl Acad. Sci. USA 116, 7371–7376 (2019).
Tilman, D. et al. The influence of functional diversity and composition on ecosystem processes. Science 277, 1300–1302 (1997).
Das, K., Prasanna, R. & Saxena, A. K. Rhizobia: a potential biocontrol agent for soilborne fungal pathogens. Folia Microbiol. 62, 425–435 (2017).
Reinhart, K. O., Tytgat, T., Van der Putten, W. H. & Clay, K. Virulence of soil-borne pathogens and invasion by Prunus serotina. New Phytol. 186, 484–495 (2010).
Hannula, S. E. et al. Shifts in rhizosphere fungal community during secondary succession following abandonment from agriculture. ISME J. 11, 2294–2304 (2017).
Heinen, R. et al. Plant community composition steers grassland vegetation via soil legacy effects. Ecol. Lett. 23, 973–982 (2020).
Forero, L. E., Grenzer, J., Heinze, J., Schittko, C. & Kulmatiski, A. Greenhouse- and field-measured plant–soil feedbacks are not correlated. Front. Environ. Sci. 7, 184 (2019).
Bagchi, R. et al. Pathogens and insect herbivores drive rainforest plant diversity and composition. Nature 506, 85–88 (2014).
Parker, I. M. et al. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520, 542–544 (2015).
Comita, L. S., Muller-Landau, H. C., Aguilar, S. & Hubbell, S. P. Asymmetric density dependence shapes species abundances in a tropical tree community. Science 329, 330–332 (2010).
Johnson, D. J., Beaulieu, W. T., Bever, J. D. & Clay, K. Conspecific negative density dependence and forest diversity. Science 336, 904–907 (2012).
Bellemain, E. et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10, 189 (2010).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Koljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277 (2013).
Abarenkov, K. et al. The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol. 186, 281–285 (2010).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2021).
Pernilla Brinkman, E., Van der Putten, W. H., Bakker, E.-J. & Verhoeven, K. J. F. Plant–soil feedback: experimental approaches, statistical analyses and ecological interpretations. J. Ecol. 98, 1063–1073 (2010).
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D. & R Core Team. nlme: linear and nonlinear mixed effects models. R package version 3.1-117 (2014); https://cran.r-project.org/web/packages/nlme/
Weiss, S. et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5, 27 (2017).
Oksanen, J. et al. The Vegan package. Community Ecol. 10, 631–637 (2007).
van Kleunen, M. et al. Economic use of plants is key to their naturalization success. Nat. Commun. 11, 3201 (2020).
Wilschut, R. A. & van Kleunen, M. Conspecific and heterospecific plant-soil feedback data and root trait measurements of 18 annual and perennial plant species. figshare https://doi.org/10.6084/m9.figshare.22740974 (2023).
Acknowledgements
We acknowledge Z. Zhang, E. Hannula, S. Geisen, R. Reuter and M. Stift for advice on statistical, molecular and root trait analyses, Q. Yang for the phylogenetic data and O. Ficht, M. Fuchs, H. Vahlenkamp, B. Speißer, B. Rüter, P. Kukofka, T. Voortman, N. Buchenau and student helpers from the University of Konstanz for practical assistance. Moreover, we acknowledge the University of Konstanz Sequencing Analysis Core Facility for assistance in analysing the fungal ITS2 sequencing data, and three reviewers for their valuable comments on the paper. R.A.W. acknowledges funding from the Wageningen Graduate Schools (WGS Postdoc Talent Grant to R.A.W.).
Author information
Authors and Affiliations
Contributions
R.A.W. and M.v.K. designed the study. R.A.W. and E.M. performed, respectively, the greenhouse experiments and molecular laboratory work. B.C.C.H. processed the raw sequencing data. Data analyses were done by R.A.W. with inputs from M.v.K. The paper was written by R.A.W. with considerable inputs from M.v.K. and was reviewed by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Brenda Casper and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Plant biomass in the conditioning phase does not explain average plant-soil-feedback responses.
Both panels show the average plant-soil-feedback response (Ln(biomassconditioned/biomasscontrol) of 18 responding plant species in relation to the average plant biomass (Ln-transformed) in pooled conditioning phase soils (see Methods). (A) Average feedback responses to all conditioned soils (N = 90; 18 conditioning species × 5 independent biological replicates). (B) Average feedback responses to all plant species, except legumes (N = 70; 14 conditioning species × 5 independent biological replicates). Results of two-sided Pearson’s correlation tests between conditioning phase biomass and average feedback responses are shown.
Extended Data Fig. 2 Root-trait variation does not predict soil-conditioning effects.
Species-level soil conditioning effects ln(biomassconditioned/biomasscontrol), averaged across 18 responding plant species, were not correlated with specific root length (A; mm/g, log-transformed), marginally significantly correlated with relative root weight (B), and not significantly correlated with average root diameter (C; mm × 10). Average trait values were obtained from a separate root-trait experiment (see Methods). Results of two-sided Pearson’s correlation tests between trait average and average feedback responses in conditioned soils are shown. In panel B, trend line and shading represent the correlation coefficient (±95% CI) between relative root weight and average PSF effect.
Extended Data Fig. 3 Fungal communities become more dissimilar with increasing phylogenetic distance.
Average pairwise Bray-Curtis dissimilarities of complete fungal communities among all 18 conditioning plant species (A), among the nine annual species (B) and among the nine perennial species (C) correlate to pairwise phylogenetic distances (ln transformed; see Methods). Results of Mantel tests between pairwise phylogenetic distances and pairwise Bray-Curtis dissimilarities are shown in each panel, while trend lines and shading represent correlation coefficients (±95% CI’s).
Extended Data Fig. 4 Putative fungal pathogen-community composition and soil-conditioning effects.
(A) NMDS-ordination showing Bray-Curtis dissimilarity-based composition of putative fungal pathogen communities, based on fungal amplicon sequence variant (ASV) abundances. PERMANOVA analysis revealed significant variation in of putative fungal pathogen community composition among conditioning plant species (see methods; full species names are listed in Supplementary Table 1). (B) Average plant-soil-feedback responses to individual conditioned soils marginally significantly vary with putative fungal pathogen community composition (NMDS-axis 2), as indicated by a linear mixed effect model and log-likelihood tests (see methods and Supplementary Table 7). In panel B, trend line and shading represent the predicted linear relationship (±95% CI) between NMDS2 and average PSF response.
Extended Data Fig. 5 Arbuscular mycorrhizal fungal community composition.
NMDS-ordination showing Bray-Curtis dissimilarity-based composition of arbuscular mycorrhizal fungal communities, based on of fungal amplicon sequence variant (ASV) abundances. PERMANOVA analysis revealed significant variation in composition of arbuscular mycorrhizal fungal communities among conditioning plant species (see methods; full species names are listed in Supplementary Table 1).
Extended Data Fig. 6 Total pathogen abundances explain soil-conditioning effects.
(A) Species-level variation in logit-transformed total pathogen abundances (means ± SEM, N = 5 independent biological replicates per plant species; full species names are listed in Supplementary Table 1). Abundances are based on all ASVs assigned as putative pathogens. (B) Average plant-soil-feedback responses to individual conditioned soils significantly vary with total pathogen abundances. In both panels, Log-likelihood ratio test results are based on linear mixed effect model (see methods and Supplementary Tables 11 & 12). In panel B, trend line and shading represent the predicted linear relationship (±95% CI) between pathogen abundance and average PSF response.
Extended Data Fig. 7 Relative abundances of generalist and specialist putative pathogens and arbuscular mycorrhizal fungi in soils conditioned by 18 annual and perennial plant species.
Fungal ASVs were manually assigned as generalists and specialists based on their occurrence in at least 2/3 or maximally 1/3 of the plant species in this study (full species names are listed in Supplementary Table 1). Relative abundances were calculated based on untransformed read counts.
Extended Data Fig. 8 Specialist-pathogen accumulation in soils of annual and perennial plant species.
Dots and bars represent means ± SEM, calculated based on average logit-transformed relative putative specialist pathogen abundances in rhizosphere soil of single plant species (N = 9 plant species per plant group (annuals/perennials)).
Supplementary information
Supplementary Information
Supplementary Tables 1–15.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wilschut, R.A., Hume, B.C.C., Mamonova, E. et al. Plant–soil feedback effects on conspecific and heterospecific successors of annual and perennial Central European grassland plants are correlated. Nat. Plants 9, 1057–1066 (2023). https://doi.org/10.1038/s41477-023-01433-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01433-w