Abstract
Plant specialized metabolites modulate developmental and ecological functions and comprise many therapeutic and other high-value compounds. However, the mechanisms determining their cell-specific expression remain unknown. Here we describe the transcriptional regulatory network that underlies cell-specific biosynthesis of triterpenes in Arabidopsis thaliana root tips. Expression of thalianol and marneral biosynthesis pathway genes depends on the phytohormone jasmonate and is limited to outer tissues. We show that this is promoted by the activity of redundant bHLH-type transcription factors from two distinct clades and coactivated by homeodomain factors. Conversely, the DOF-type transcription factor DAG1 and other regulators prevent expression of the triterpene pathway genes in inner tissues. We thus show how precise expression of triterpene biosynthesis genes is determined by a robust network of transactivators, coactivators and counteracting repressors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The scRNAseq data are accessible via an online browser tool (http://bioit3.irc.ugent.be/plant-sc-atlas/) and raw data are deposited at NCBI with GEO numbers GSE179820 and GSE212826 for the mock- and JA-treated root tips, respectively. All other data generated for this study are included either in the main paper, Extended Data, or the Supplementary Information. Material requests should be directed to the corresponding author. Published data for TF motif mapping were retrieved from CisBP 2.00 (downloaded in December 2019: http://cisbp.ccbr.utoronto.ca/) and JASPAR2020 (https://jaspar.genereg.net/). The regulatory regions used for motif mapping were downloaded from PLAZA Dicots 4.5 (https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v4_5_dicots/). Source data are provided with this paper.
References
Erb, M. & Kliebenstein, D. J. Plant secondary metabolites as defenses, regulators, and primary metabolites: the blurred functional trichotomy. Plant Physiol. 184, 39–52 (2020).
Lacchini, E. & Goossens, A. Combinatorial control of plant specialized metabolism: mechanisms, functions, and consequences. Annu. Rev. Cell Dev. Biol. 36, 291–313 (2020).
Colinas, M. & Goossens, A. Combinatorial transcriptional control of plant specialized metabolism. Trends Plant Sci. 23, 324–336 (2018).
Wasternack, C. & Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites – pathways, transcription factors and applied aspects – a brief review. New Biotechnol. 48, 1–11 (2019).
Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 148, 63–106 (2015).
Thimmappa, R., Geisler, K., Louveau, T., O’Maille, P. & Osbourn, A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65, 225–257 (2014).
Huang, A. C. et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364, eaau6389 (2019).
Pichersky, E. & Raguso, R. A. Why do plants produce so many terpenoid compounds? New Phytol. 220, 692–702 (2018).
Sohrabi, R. et al. In planta variation of volatile biosynthesis: an alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell 27, 874–890 (2015).
Tholl, D. & Lee, S. Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book 9, e0143 (2011).
Field, B. et al. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl Acad. Sci. USA 108, 16116–16121 (2011).
Field, B. & Osbourn, A. E. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science 320, 543–547 (2008).
Nützmann, H.-W. et al. Active and repressed biosynthetic gene clusters have spatially distinct chromosome states. Proc. Natl Acad. Sci. USA 117, 13800–13809 (2020).
Nützmann, H.-W., Huang, A. & Osbourn, A. Plant metabolic clusters – from genetics to genomics. New Phytol. 211, 771–789 (2016).
Bai, Y. et al. Modulation of Arabidopsis root growth by specialized triterpenes. New Phytol. 230, 228–243 (2021).
Goossens, J., Fernández-Calvo, P., Schweizer, F. & Goossens, A. Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91, 673–689 (2016).
Chini, A., Gimenez-Ibanez, S., Goossens, A. & Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156 (2016).
Pauwels, L. et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Fernández-Calvo, P. et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715 (2011).
Thines, B. et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665 (2007).
Fonseca, S. et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350 (2009).
Sheard, L. B. et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405 (2010).
Yan, Y. et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483 (2007).
Gasperini, D. et al. Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genet. 11, e1005300 (2015).
Major, I. T. et al. Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 215, 1533–1547 (2017).
Lorenzo, O., Chico, J. M., Sánchez-Serrano, J. J. & Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950 (2004).
Schweizer, F. et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132 (2013).
Birnbaum, K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960 (2003).
Zander, M. et al. Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 6, 290–302 (2020).
Mertens, J. et al. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 170, 194–210 (2016).
Van Moerkercke, A. et al. The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc. Natl Acad. Sci. USA 112, 8130–8135 (2015).
Van Moerkercke, A. et al. A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl Acad. Sci. USA 116, 23345–23356 (2019).
Goossens, J., Swinnen, G., Vanden Bossche, R., Pauwels, L. & Goossens, A. Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 206, 1229–1237 (2015).
Kemen, A. C. et al. Investigation of triterpene synthesis and regulation in oats reveals a role for β-amyrin in determining root epidermal cell patterning. Proc. Natl Acad. Sci. USA 111, 8679–8684 (2014).
Ohashi, Y. et al. Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300, 1427–1430 (2003).
Wang, S., Barron, C., Schiefelbein, J. & Chen, J. G. Distinct relationships between GLABRA2 and single‐repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185, 387–400 (2010).
Gualberti, G. et al. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14, 1253–1263 (2002).
Ramachandran, V. et al. Plant-specific Dof transcription factors VASCULAR-RELATED DOF1 and VASCULAR-RELATED DOF2 regulate vascular cell differentiation and lignin biosynthesis in Arabidopsis. Plant Mol. Biol. 104, 263–281 (2020).
Boccaccini, A. et al. The DOF protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. Mol. Plant 7, 1486–1489 (2014).
Papi, M. et al. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 14, 28–33 (2000).
Gabriele, S. et al. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61, 312–323 (2010).
Zhao, H. et al. Identification and functional validation of super-enhancers in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 119, e2215328119 (2022).
Baudry, A., Caboche, M. & Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779 (2006).
Ellis, C. & Turner, J. G. A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556 (2002).
Xie, D.-X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Decaestecker, W. et al. CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31, 2868–2887 (2019).
Vanden Bossche, R., Demedts, B., Vanderhaeghen, R. & Goossens, A. Transient expression assays in tobacco protoplasts. Methods Mol. Biol. 1011, 227–239 (2013).
Wendrich, J. R. et al. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370, eaay4970 (2020).
Yang, B. et al. Non-cell autonomous and spatiotemporal signalling from a tissue organizer orchestrates root vascular development. Nat. Plants 7, 1485–1494 (2021).
Lun, A. T., Bach, K. & Marioni, J. C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75 (2016).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019).
Lambert, S. A. et al. Similarity regression predicts evolution of transcription factor sequence specificity. Nat. Genet. 51, 981–989 (2019).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020).
Kulkarni, S. R., Vaneechoutte, D., Van de Velde, J. & Vandepoele, K. TF2Network: predicting transcription factor regulators and gene regulatory networks in Arabidopsis using publicly available binding site information. Nucleic Acids Res. 46, e31 (2017).
Frith, M. C., Li, M. C. & Weng, Z. Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 31, 3666–3668 (2003).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Cheng, C.-Y. et al. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017).
Van Bel, M. et al. PLAZA 5.0: extending the scope and power of comparative and functional genomics in plants. Nucleic Acids Res. 50, D1468–D1474 (2022).
Kulkarni, S. R., Jones, D. M. & Vandepoele, K. Enhanced maps of transcription factor binding sites improve regulatory networks learned from accessible chromatin data. Plant Physiol. 181, 412–425 (2019).
Acknowledgements
This Article was written in loving memory of A. Van Moerkercke (1979–2021). The authors thank A. Bleys for critically reading the manuscript; D. Gasperini for kindly sharing the ProMYCs:NLS-VENUS reporter lines, and P. Vittorioso for the dag1 mutant, ProDAG1:GUS and DAG1 over-expressing lines; J. R. Wendrich and T. Eekhout for assistance in the launching and analysis of the scRNAseq experiment; and S. Desmet and G. Goeminne from the VIB Metabolomics Core – Ghent for the thalianol profiling. This work was supported by the European Community’s Seventh Framework Program (FP7/2007–2013) under grant agreement 613692-TriForC and H2020 Program under grant agreement 760331-Newcotiana to A.G.; the Special Research Fund from Ghent University to A.G. and A.R. (project BOF18/GOA/013), and M.M. (project BOF20/GOA/012); the Flemish Government (AI Research program) to Y.S.; the Research Foundation Flanders with research project grants to A.G. (G004515N and G008417N) and a postdoctoral fellowship to P.F.-C.; a Swiss National Science Foundation postdoctoral fellowship (P300PA_177831) to M.C.; and a China Scholarship Council PhD scholarship to Y.B. A.O. acknowledges funding support from the John Innes Foundation and the BBSRC Institute Strategic Program Grant ‘Molecules from Nature – Products and Pathways’ (BBS/E/J/000PR9790).
Author information
Authors and Affiliations
Contributions
A.G., T.H.N., A.V.M., P.F.-C. and A.R. conceptualized the project. T.H.N., L.T., M.M., T.D., M.C., K. Verstaen, G.V.I., Y.S., B.D.R., K. Vandepoele, H.-W.N., A.O., A.R. and A.G. developed the methodology. T.H.N., Y.B., A.V.M., P.F.-C., L.T., T.D., M.C., G.V.I., A.R. and A.G. conducted the investigations. T.H.N., L.T., A.R., K. Verstaen and T.D. performed visualization. A.G, Y.S. and A.O. acquired funding. A.G. and A.R. administered the project. A.G., A.R., B.D.R., K. Vandepoele and Y.S. supervised the project. A.V.M., A.R. and A.G. wrote the original draft. T.H.N. and B.D.R. reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Yang-Dong Guo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1
Violin plots showing cell-specific expression of the thalianol biosynthesis genes under mock and JA treatments.
Extended Data Fig. 2
Violin plots showing cell-specific expression of the marneral biosynthesis genes under mock and JA treatments.
Extended Data Fig. 3 MYC2 expression is induced by protoplasting in root tips.
a, Violin plots showing cell-specific expression of MYC2, MYC3 and MYC4 upon mock and JA treatments. b, RT-qPCR expression analysis of intact roots tips and root tip protoplasts upon mock and JA treatments. Values on the Y-axis represent fold-induction compared to mock-treated intact roots (set to 1). The error bars designate the SE of the mean (n = 3 biologically independent samples). Statistical significance was determined using the Student’s t-test (*P < 0.05, **P < 0.005, ***P < 0.0005).
Extended Data Fig. 4 Heatmap of THAS single-cell co-expression analysis.
Top 100 genes co-expressed with THAS under JA treatment across all root tip tissue types. Expression values as log fold change of JA compared to mock conditions are shown for each of the separate tissue types.
Extended Data Fig. 5 The expression of bHLH clade IVa genes is induced by JA in a COI1-dependent manner.
a, Violin plots showing cell-specific expression of bHLH18 and bHLH25 upon mock and JA treatments. b, Expression profile of ProbHLH19:GFP-GUS in wt root tips grown on mock or 50 µM JA for 24 h. Scale bars = 20 µm. At least 3 seedling roots were observed with similar results. c, RT-qPCR expression analysis of bHLH clade IV genes upon mock and JA treatments in the Col-0 wt and coi1-1 backgrounds. The error bars designate the SE of the mean (n = 3 biologically independent samples). Values on the Y-axis represent fold-induction compared to mock-treated wt (set to 1). Statistical significance was determined using the Student’s t-test (*P < 0.05, **P < 0.005, ***P < 0.0005).
Extended Data Fig. 6 Expression of bHLH clade IVa transcription factors is induced by MYCs.
a, RT-qPCR expression analysis of bHLH clade IV genes upon mock and JA treatments in the Col-0 wt and mycT backgrounds. The error bars designate the SE of the mean (n = 3 biologically independent samples). Values on the Y-axis represent fold-induction compared to mock-treated wt (set to 1). Statistical significance was determined using the Student’s t-test (*P < 0.05, **P < 0.005, ***P < 0.0005). b, Transactivation in N. tabacum protoplasts of the ProbHLH19 and ProbHLH20 fused to the fLUC reporter and cotransfected with either GUS, MYC2, MYC2D105N, bHLH19 or bHLH25. Values on the Y-axis are normalized fold-changes relative to protoplasts cotransfected with the reporter constructs and a pCaMV35S:GUS (GUS) control plasmid (set to 1). The error bars designate the SE of the mean (n = 8 biologically independent samples). Statistical significance was determined using the Student’s t-test (*P < 0.05, **P < 0.005, ***P < 0.0005).
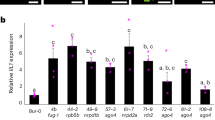
Extended Data Fig. 7 Violin plots showing cell-specific expression of candidate root triterpene transcriptional regulators under mock- and JA-treated conditions.
Only genes from the HDG- and DOF-type families that show expression in our scRNAseq dataset are represented.
Extended Data Fig. 8 Homeodomain glabrous proteins coactivate the transcription of the THAS promoter.
Transactivation in N. tabacum protoplasts transfected with ProTHAS fused to the fLUC reporter, and cotransfected with combinations of GL2, HDG2, HDG5, MYC2D105N, bHLH19 or/and bHLH20. Values on the Y-axis are normalized fold-changes relative to protoplasts cotransfected with the reporter constructs and a pCaMV35S:GUS (GUS) control plasmid (set to 1). The error bars designate the SE of the mean (n = 8 biologically independent samples). Statistical significance was determined using the Student’s t-test (*P < 0.05, **P < 0.005, ***P < 0.0005).
Extended Data Fig. 9 vDOF1 proteins do not modulate transcription of the THAS promoter.
Transactivation in transfected N. tabacum protoplasts of the ProTHAS fused to the fLUC reporter, and cotransfected with combinations of vDOF1, MYC2D105N, bHLH19 or/and bHLH20. Values on the Y-axis are normalized fold-changes relative to protoplasts co-transfected with the reporter constructs and a pCaMV35S:GUS (GUS) control plasmid (set to 1). The error bars designate the SE of the mean (n = 8 biologically independent samples). Statistical significance was determined using the Student’s t-test (NS, Non-significant; ***P < 0.0005).
Extended Data Fig. 10
Model for the regulatory network that drives spatiotemporal expression of thalianol and marneral biosynthesis genes in Arabidopsis root tips.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Tables 1–5.
Supplementary Data
Source data for supplementary figures.
Source data
Source Data Figs. 1–4 and Extended Data Figs. 3, 5, 6, 8 and 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nguyen, T.H., Thiers, L., Van Moerkercke, A. et al. A redundant transcription factor network steers spatiotemporal Arabidopsis triterpene synthesis. Nat. Plants 9, 926–937 (2023). https://doi.org/10.1038/s41477-023-01419-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01419-8