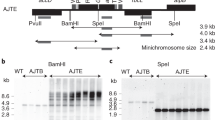
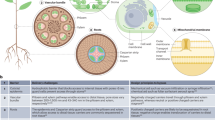
Abstract
Engineering the plastid genome based on homologous recombination is well developed in a few model species. Homologous recombination is also the rule in mitochondria, but transformation of the mitochondrial genome has not been realized in the absence of selective markers. The application of transcription activator-like (TAL) effector-based tools brought about a dramatic change because they can be deployed from nuclear genes and targeted to plastids or mitochondria by an N-terminal targeting sequence. Recognition of the target site in the organellar genomes is ensured by the modular assembly of TALE repeats. In this paper, I review the applications of TAL effector nucleases and TAL effector cytidine deaminases for gene deletion, base editing and mutagenesis in plastids and mitochondria. I also review emerging technologies such as post-transcriptional RNA modification to regulate gene expression, Agrobacterium- and nanoparticle-mediated organellar genome transformation, and self-replicating organellar vectors as production platforms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Gehrke, F., Schindele, A. & Puchta, H. Non-homologous end joining as key to CRISPR/Cas-mediated plant chromosome engineering. Plant Physiol. 188, 1769–1779 (2022).
He, Y., Mudgett, M. & Zhao, Y. Advances in gene editing without residual transgenes in plants. Plant Physiol. 188, 1757–1761 (2022).
Gurdon, C., Svab, Z., Feng, Y., Kumar, D. & Maliga, P. Cell-to-cell movement of mitochondria in plants. Proc. Natl Acad. Sci. USA 113, 3395–3400 (2016).
Sanchez-Puerta, M. V., Zubko, M. K. & Palmer, J. D. Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 206, 381–396 (2015).
Kwon, T., Huq, E. & Herrin, D. L. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc. Natl Acad. Sci. USA 107, 13954–13959 (2010).
Kazama, T. et al. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat. Plants 5, 722–730 (2019).
Arimura, S. Effects of mitoTALENs-directed double-strand breaks on plant mitochondrial genomes comment. Genes https://doi.org/10.3390/genes12020153 (2021).
Svab, Z., Hajdukiewicz, P. & Maliga, P. Stable transformation of plastids in higher plants. Proc. Natl Acad. Sci. USA 87, 8526–8530 (1990).
Bock, R. Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66, 211–241 (2015).
Maliga, P. in Genomics of Chloroplasts and Mitochondria Vol. 35 (eds Bock, R. & Knoop, V.) 393–414 (Springer, 2012).
Klaus, S. M. J., Huang, F. C., Golds, T. J. & Koop, H. U. Generation of marker-free plastid transformants using a transiently cointegrated selection gene. Nat. Biotechnol. 22, 225–229 (2004).
Kode, V., Mudd, E. A., Iamtham, S. & Day, A. Isolation of precise plastid deletion mutants by homology-based excision: a resource for site-directed mutagenesis, multi-gene changes and high-throughput plastid transformation. Plant J. 46, 901–909 (2006).
Martin Avila, E., Gisby, M. F. & Day, A. Seamless editing of the chloroplast genome in plants. BMC Plant Biol. 16, 168 (2016).
Maliga, P. & Walker, J. M. (eds) Methods in Molecular Biology 2nd edn Vol. 2317 (Springer, 2021).
Wilson, D. N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12, 35–48 (2014).
Wirmer, J. & Westhof, E. Molecular contacts between antibiotics and the 30S ribosomal particle. Methods Enzymol. 415, 180–202 (2006).
Yu, Q., Lutz, K. A. & Maliga, P. Efficient plastid transformation in Arabidopsis. Plant Physiol. 175, 186–193 (2017).
Zubko, M. K. & Day, A. Stable albinism induced without mutagenesis: a model for ribosome-free plastid inheritance. Plant J. 15, 265–271 (1998).
Parker, N., Wang, Y. & Meinke, D. Natural variation in sensitivity to a loss of chloroplast translation in Arabidopsis. Plant Physiol. 166, 2013–2027 (2014).
Yu, Q., LaManna, L., Kelly, M. E., Lutz, K. A. & Maliga, P. New tools for engineering the Arabidopsis plastid genome. Plant Physiol. 181, 394–398 (2019).
Ruf, S. et al. High-efficiency generation of fertile transplastomic Arabidopsis plants. Nat. Plants 5, 282–289 (2019).
Silhavy, D. & Maliga, P. Mapping of the promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet. 33, 340–344 (1998).
Khan, M. S. & Maliga, P. Fluorescent antibiotic resistance marker to track plastid transformation in higher plants. Nat. Biotechnol. 17, 910–915 (1999).
Lee, S. M. et al. Plastid transformation in the monocotyledonous cereal crop, rice (Oryza sativa) and transmission of transgenes to their progeny. Mol. Cells 21, 401–410 (2006).
Li, R. Q. et al. High-efficiency plastome base editing in rice with TAL cytosine deaminase. Mol. Plant 14, 1412–1414 (2021).
Prikryl, J., Rojas, M., Schuster, G. & Barkan, A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA 108, 415–420 (2011).
Pfalz, J., Bayraktar, O. A., Prikryl, J. & Barkan, A. Site-specific binding of a PPR protein defines and stabilizes 5’ and 3’ mRNA termini in chloroplasts. EMBO J. 28, 2042–2052 (2009).
Yu, Q., Barkan, A. & Maliga, P. Engineered RNA-binding protein for transgene activation in non-green plastids. Nat. Plants 5, 486–490 (2019).
Barkan, A. et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 (2012).
Gordon, K. H. & Waterhouse, P. M. RNAi for insect-proof plants. Nat. Biotechnol. 25, 1231–1232 (2007).
Zhang, J., Khan, S. A., Heckel, D. G. & Bock, R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 35, 871–882 (2017).
Bally, J. et al. Improved insect-proofing: expressing double-stranded RNA in chloroplasts. Pest Manage. Sci. 74, 1751–1758 (2018).
Zhang, J. et al. Pest control. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 347, 991–994 (2015).
Bally, J. et al. In-plant protection against Helicoverpa armigera by production of long hpRNA in chloroplasts. Front. Plant Sci. 7, 1453 (2016).
Jin, S., Singh, N. D., Li, L., Zhang, X. & Daniell, H. Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa zea larval development and pupation. Plant Biotechnol. J. 13, 435–446 (2015).
Wu, M. et al. Efficient control of western flower thrips by plastid-mediated RNA interference. Proc. Natl Acad. Sci. USA (in the press).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Sakuma, T. et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 3, 3379 (2013).
Kang, B. C. et al. Chloroplast and mitochondrial DNA editing in plants. Nat. Plants 7, 899–905 (2021).
Lee, L. Y. & Gelvin, S. B. T-DNA binary vectors and systems. Plant Physiol. 146, 325–332 (2008).
Komori, T. et al. Current status of binary vectors and superbinary vectors. Plant Physiol. 145, 1155–1160 (2007).
Joung, J. K. & Sander, J. D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 (2013).
Beurdeley, M. et al. Compact designer TALENs for efficient genome engineering. Nat. Commun. 4, 1762 (2013).
Arimura, S. et al. Targeted gene disruption of ATP synthases 6-1 and 6-2 in the mitochondrial genome of Arabidopsis thaliana by mitoTALENs. Plant J. 104, 1459–1471 (2020).
Mok, B. Y. et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020).
Hanson, M. R. & Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16, S154–S169 (2004).
Chen, L. & Liu, Y. G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606 (2014).
Omukai, S., Arimura, S., Toriyama, K. & Kazama, T. Disruption of mitochondrial open reading frame 352 partially restores pollen development in cytoplasmic male sterile rice. Plant Physiol. 187, 236–246 (2021).
Takatsuka, A., Kazama, T., Arimura, S. & Toriyama, K. TALEN-mediated depletion of the mitochondrial gene orf312 proves that it is a Tadukan-type cytoplasmic male sterility-causative gene in rice. Plant J. https://doi.org/10.1111/tpj.15715 (2022).
Kuwabara, K., Arimura, S., Shirasawa, K. & Ariizumi, T. orf137 triggers cytoplasmic male sterility in tomato. Plant Physiol. 189, 465–468 (2022).
Forner, J. et al. Targeted introduction of heritable point mutations into the plant mitochondrial genome. Nat. Plants 8, 245 (2022).
Aouida, M., Piatek, M. J., Bangarusamy, D. K. & Mahfouz, M. M. Activities and specificities of homodimeric TALENs in Saccharomyces cerevisiae. Curr. Genet. 60, 61–74 (2014).
Sanders, K. L., Catto, L. E., Bellamy, S. R. & Halford, S. E. Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands. Nucleic Acids Res. 37, 2105–2115 (2009).
Preuten, T. et al. Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 64, 948–959 (2010).
Giege, P. & Brennicke, A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA 96, 15324–15329 (1999).
Nakazato, I. et al. Targeted base editing in the mitochondrial genome of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 119, e2121177119 (2022).
Fromm, H., Galun, E. & Edelman, M. A novel site for streptomycin resistance in the ‘530 loop’ of chloroplast 16S ribosomal RNA. Plant Mol. Biol. 12, 499–505 (1989).
Nakazato, I. et al. Targeted base editing in the plastid genome of Arabidopsis thaliana. Nat. Plants 7, 906–913 (2021).
Hoch, B., Maier, R. M., Appel, K., Igloi, G. L. & Kössel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353, 178–180 (1991).
Kahlau, S., Aspinall, S., Gray, J. C. & Bock, R. Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J. Mol. Evol. 63, 194–207 (2006).
Tillich, M. et al. Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 43, 708–715 (2005).
Ruwe, H., Castandet, B., Schmitz-Linneweber, C. & Stern, D. B. Arabidopsis chloroplast quantitative editotype. FEBS Lett. 587, 1429–1433 (2013).
Small, I. D., Schallenberg-Rudinger, M., Takenaka, M., Mireau, H. & Ostersetzer-Biran, O. Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 101, 1040–1056 (2020).
Barkan, A. & Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 (2014).
Sun, T., Bentolila, S. & Hanson, M. R. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 21, 962–973 (2016).
Oldenkott, B. et al. One C-to-U RNA editing site and two independently evolved editing factors: testing reciprocal complementation with DYW-type PPR proteins from the moss Physcomitrium (Physcomitrella) patens and the flowering plants Macadamia integrifolia and Arabidopsis. Plant Cell 32, 2997–3018 (2020).
Royan, S. et al. A synthetic RNA editing factor edits its target site in chloroplasts and bacteria. Commun. Biol. https://doi.org/10.1038/s42003-021-02062-9 (2021).
Bernath-Levin, K. et al. Cofactor-independent RNA editing by a synthetic S-type PPR protein. Synth. Biol. https://doi.org/10.1093/synbio/ysab034 (2022).
Arroyo-Olarte, R. D., Rodriguez, R. B. & Morales-Rios, E. Genome editing in bacteria: CRISPR-Cas and beyond. Microorganisms https://doi.org/10.3390/microorganisms9040844 (2021).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Tong, Y., Jorgensen, T. S., Whitford, C. M., Weber, T. & Lee, S. Y. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat. Commun. 12, 5206 (2021).
Niazi, A. K. et al. Targeting nucleic acids into mitochondria: progress and prospects. Mitochondrion 13, 548–558 (2013).
Tarasenko, T. A. et al. Plant mitochondria import DNA via alternative membrane complexes involving various VDAC isoforms. Mitochondrion 60, 43–58 (2021).
Handa, H. Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions. Mitochondrion 8, 15–25 (2008).
Matsuoka, A. & Maliga, P. Progress in reengineering Agrobacterium T-DNA delivery to chloroplasts. Plant Physiol. 186, 215–220 (2021).
Santana, I., Wu, H. H., Hu, P. G. & Giraldo, J. P. Targeted delivery of nanomaterials with chemical cargoes in plants enabled by a biorecognition motif. Nat. Commun. https://doi.org/10.1038/s41467-020-15731-w (2020).
Yoshizumi, T., Oikawa, K., Chuah, J. A., Kodama, Y. & Numata, K. Selective gene delivery for integrating exogenous DNA into plastid and mitochondria! genomes using peptide-DNA complexes. Biomacromolecules 19, 1582–1591 (2018).
Thagun, C. et al. Non-transgenic gene modulation via spray delivery of nucleic acid/peptide complexes into plant nuclei and chloroplasts. ACS Nano 16, 3506–3521 (2022).
Jakubiec, A. et al. Replicating minichromosomes as a new tool for plastid genome engineering. Nat. Plants 7, 932 (2021).
Jensen, P. E. & Scharff, L. B. Engineering of plastids to optimize the production of high-value metabolites and proteins. Curr. Opin. Biotechnol. 59, 8–15 (2019).
Li, S., Chang, L. & Zhang, J. Advancing organelle genome transformation and editing for crop improvement. Plant Commun. 2, 100141 (2021).
Hanson, M. R., Lin, M. T., Carmo-Silva, A. E. & Parry, M. A. Towards engineering carboxysomes into C3 plants. Plant J. 87, 38–50 (2016).
Martin-Avila, E. et al. Modifying plant photosynthesis and growth via simultaneous chloroplast transformation of rubisco large and small subunits. Plant Cell 32, 2898–2916 (2020).
Svab, Z. & Maliga, P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA 90, 913–917 (1993).
Ruf, S., Hermann, M., Berger, I. J., Carrer, H. & Bock, R. Stable genetic transformation of tomato plastids: foreign protein expression in fruit. Nat. Biotechnol. 19, 870–875 (2001).
Dufourmantel, N. et al. Generation of fertile transplastomic soybean. Plant Mol. Biol. 55, 479–489 (2004).
Cho, S. H., Chung, Y. S., Cho, S. K., Rim, Y. W. & Shin, J. S. Particle bombardment mediated transformation and GFP expression in the moss Physcomitrella patens. Mol. Cells 9, 14–19 (1999).
Zubko, M. K., Zubko, E. I., van Zuilen, K., Mayer, P. & Day, A. Stable transformation of petunia plastids. Transgenic Res. 13, 523–530 (2004).
Kanamoto, H. et al. Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res 15, 205–217 (2006).
Ruhlman, T., Verma, D., Samson, N. & Daniell, H. The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 152, 2088–2104 (2010).
Okumura, S. et al. Transformation of poplar (Populus alba) plastids and expression of foreign proteins in tree chloroplasts. Transgenic Res. 15, 637–646 (2006).
Liu, C. W., Lin, C. C., Chen, J. J. & Tseng, M. J. Stable chloroplast transformation in cabbage (Brassica oleracea L. var. capitata L.) by particle bombardment. Plant Cell Rep. 26, 1733–1744 (2007).
De Marchis, F., Wang, Y., Stevanato, P., Arcioni, S. & Bellucci, M. Genetic transformation of the sugar beet plastome. Transgenic Res. 18, 17–30 (2009).
Valkov, V. T. et al. High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5’ and 3’ regulatory sequences. Transgenic Res. 20, 137–151 (2010).
Davarpanah, S. J. et al. Stable plastid transformation in Nicotiana benthamiana. J. Plant Biol. 52, 244–250 (2009).
Boehm, C. R., Ueda, M., Nishimura, Y., Shikanai, T. & Haseloff, J. A cyan fluorescent reporter expressed from the chloroplast genome of Marchantia polymorpha. Plant Cell Physiol. 57, 291–299 (2016).
Lutz, K. A. & Maliga, P. Construction of marker-free transplastomic plants. Curr. Opin. Biotechnol. 18, 107–114 (2007).
Acknowledgements
P.M. thanks C. Best for critical reading of the manuscript. Original research cited was supported by Research Grants from the National Science Foundation MCB 1716102 and National Science Foundation IOS 2037155 to P.M., and USDA NIFA Foundational Program Award No. 2014-67013-21600 to A. Barkan and P.M.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Peer review
Peer review information
Nature Plants thanks Anil Day, Shin-ichi Arimura and Spencer Whitney for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Maliga, P. Engineering the plastid and mitochondrial genomes of flowering plants. Nat. Plants 8, 996–1006 (2022). https://doi.org/10.1038/s41477-022-01227-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01227-6
This article is cited by
-
Base editing of organellar DNA with programmable deaminases
Nature Reviews Molecular Cell Biology (2024)
-
TALE-based organellar genome editing and gene expression in plants
Plant Cell Reports (2024)
-
Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications
Molecular Biomedicine (2023)
-
Targeted knockout of a conserved plant mitochondrial gene by genome editing
Nature Plants (2023)