Abstract
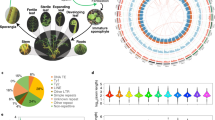
Euphyllophytes encompass almost all extant plants, including two sister clades, ferns and seed plants. Decoding genomes of ferns is the key to deep insight into the origin of euphyllophytes and the evolution of seed plants. Here we report a chromosome-level genome assembly of Adiantum capillus-veneris L., a model homosporous fern. This fern genome comprises 30 pseudochromosomes with a size of 4.8-gigabase and a contig N50 length of 16.22 Mb. Gene co-expression network analysis uncovered that homospore development in ferns has relatively high genetic similarities with that of the pollen in seed plants. Analysing fern defence response expands understanding of evolution and diversity in endogenous bioactive jasmonates in plants. Moreover, comparing fern genomes with those of other land plants reveals changes in gene families important for the evolutionary novelties within the euphyllophyte clade. These results lay a foundation for studies on fern genome evolution and function, as well as the origin and evolution of euphyllophytes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The A. capillus-veneris genome assembly, genome annotation and all the raw sequencing data have been deposited at NCBI, under the BioProject accession number PRJNA593372 (genome assembly and annotation) and PRJNA593361 (transcriptome raw sequence data). The CDS and peptide files are available from https://figshare.com/s/47be9fe90124b22d3c0e. The referenced dataset for Arabidopsis LEC1 co-expression analysis is under the accessions: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12404 (DOI: 10.1073/pnas.1707957114). Source data are provided with this paper.
Code availability
All custom codes are available for research purposes from the corresponding authors upon request.
References
Sessa, E. B. & Der, J. P. in Advances in Botanical Research (ed Rsening, S. A.) 215–254 (Academic Press, 2016).
The Pteridophyte Phylogeny Group. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 54, 563–603 (2016).
Pryer, K. M. et al. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409, 618–622 (2001).
Chang, C., Bowman, J. L. & Meyerowitz, E. M. Field guide to plant model systems. Cell 167, 325–339 (2016).
Wolf, P. G. et al. An exploration into fern genome space. Genome Biol. Evol. 7, 2533–2544 (2015).
Szövényi, P., Gunadi, A. & Li, F. W. Charting the genomic landscape of seed-free plants. Nat. Plants 7, 554–565 (2021).
Clark, J. et al. Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 210, 1072–1082 (2016).
Li, F. W. et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants 4, 460–472 (2018).
Huang, X. et al. The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence. Nat. Plants 8, 500–512 (2022).
Sessa, E. B. et al. Between two fern genomes. Gigascience 3, 15 (2014).
Li, X., Fang, Y. H., Yang, J., Bai, S. N. & Rao, G. Y. Overview of the morphology, anatomy, and ontogeny of Adiantum capillus-veneris: an experimental system to study the development of ferns. J. Syst. Evol. 51, 499–510 (2013).
Tsuboi, H., Suetsugu, N., Kawai-Toyooka, H. & Wada, M. Phototropins and neochrome1 mediate nuclear movement in the fern Adiantum capillus-veneris. Plant Cell Physiol. 48, 892–896 (2007).
Wada, M. Chloroplast and nuclear photorelocation movements. Proc. Jpn. Acad. B 92, 387–411 (2016).
Shen, H. et al. Large-scale phylogenomic analysis resolves a backbone phylogeny in ferns. Gigascience 7, 1–11 (2018).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Leebens-Mack, J. H. et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019).
Huang, C. H., Qi, X., Chen, D., Qi, J. & Ma, H. Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. J. Integr. Plant Biol. 62, 433–455 (2020).
Sensalari, C., Maere, S. & Lohaus, R. ksrates: positioning whole-genome duplications relative to speciation events in KS distributions. Bioinformatics 38, 530–532 (2022).
Sato, N. & Furuya, M. The composition of lipids and fatty acids determined at various stages of haploid and diploid generations in the fern Adiantum capillus-veneris. Physiol. Plant. 62, 139–147 (1984).
Alamillo, J. M. & Bartels, D. Effects of desiccation on photosynthesis pigments and the ELIP-like dsp 22 protein complexes in the resurrection plant Craterostigma plantagineum. Plant Sci. 160, 1161–1170 (2001).
Manfre, A. J., LaHatte, G. A., Climer, C. R. & Marcotte, W. R. Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana Mutant atem6-1. Plant Cell Physiol. 50, 243–253 (2008).
Alves, M. S., Fontes, E. P. B. & Fietto, L. G. EARLY RESPONSIVE to DEHYDRATION 15, a new transcription factor that integrates stress signaling pathways. Plant Signal. Behav. 6, 1993–1996 (2011).
Kagaya, Y. et al. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46, 399–406 (2005).
West, M. A. L. et al. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6, 1731–1745 (1994).
Xie, Z. et al. Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol. Biol. Evol. 25, 1581–1592 (2008).
Sreenivasulu, N. & Wobus, U. Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64, 189–217 (2013).
Braybrook, S. A. & Harada, J. J. LECs go crazy in embryo development. Trends Plant Sci. 13, 624–630 (2008).
Pelletier, J. M. et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl Acad. Sci. USA 114, E6710–E6719 (2017).
Mertens, J., Aliyu, H. & Cowan, D. A. LEA proteins and the evolution of the WHy domain. Appl. Environ. Microbiol. 84, e00539-18 (2018).
Olvera-Carrillo, Y., Luis Reyes, J. & Covarrubias, A. A. Late embryogenesis abundant proteins: versatile players in the plant adaptation to water limiting environments. Plant Signal. Behav. 6, 586–589 (2011).
Fatihi, A. et al. Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Sci. 250, 198–204 (2016).
Kirkbride, R. C., Fischer, R. L. & Harada, J. J. LEAFY COTYLEDON1, a key regulator of seed development, is expressed in vegetative and sexual propagules of Selaginella moellendorffii. PLoS ONE 8, e67971 (2013).
Markham, K., Chalk, T. & Stewart, C. N. Jr. Evaluation of fern and moss protein-based defenses against phytophagous insects. Int. J. Plant Sci. 167, 111–117 (2006).
Hendrix, S. D. An evolutionary and ecological perspective of the insect fauna of ferns. Am. Nat. 115, 171–196 (1980).
Ali, M. S. & Baek, K. H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 21, 621 (2020).
Monte, I. et al. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 14, 480–488 (2018).
Scholz, J. et al. Biosynthesis of allene oxides in Physcomitrella patens. BMC Plant Biol. 12, 228 (2012).
Koo, A. J. K., Gao, X., Daniel Jones, A. & Howe, G. A. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986 (2009).
Reymond, P., Weber, H., Damond, M. & Farmer, E. E. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719 (2000).
de Vries, S. et al. Jasmonic and salicylic acid response in the fern Azolla filiculoides and its cyanobiont. Plant. Cell Environ. 41, 2530–2548 (2018).
Schüler, G. et al. Coronalon: a powerful tool in plant stress physiology. FEBS Lett. 563, 17–22 (2004).
Pratiwi, P. et al. Identification of jasmonic acid and jasmonoyl-isoleucine, and characterization of AOS, AOC, OPR and JAR1 in the model lycophyte Selaginella moellendorffii. Plant Cell Physiol. 58, 789–801 (2017).
Fan, S. et al. Limonin: a review of its pharmacology, toxicity, and pharmacokinetics. Molecules 24, 3679 (2019).
Kim, S. et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109 (2019).
Sherer, T. B. et al. Mechanism of toxicity in rotenone models of Parkinson’s disease. J. Neurosci. 23, 10756–10764 (2003).
Wang, H. & Mao, H. On the origin and evolution of plant brassinosteroid receptor kinases. J. Mol. Evol. 78, 118–129 (2014).
Caño-Delgado, A. et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular defferentiation in Arabidopsis. Development 131, 5341–5351 (2004).
Zheng, B. et al. EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat. Commun. 10, 4165 (2019).
Ceserani, T., Trofka, A., Gandotra, N. & Nelson, T. VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J. 57, 1000–1014 (2009).
Shiu, S. H. et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234 (2004).
Marchant, D. B. et al. The C-Fern (Ceratopteris richardii) genome: insights into plant genome evolution with the first partial homosporous fern genome assembly. Sci. Rep. 9, 18181 (2019).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020).
Gawel, N. J. & Jarret, R. L. A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol. Biol. Rep. 9, 262–266 (1991).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Du, H. & Liang, C. Assembly of chromosome-scale contigs by efficiently resolving repetitive sequences with long reads. Nat. Commun. 10, 5360 (2019).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Rice, A. et al. The Chromosome Counts Database (CCDB) – a community resource of plant chromosome numbers. New Phytol. 206, 19–26 (2015).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the european molecular biology open software suite. Trends Genet. 16, 276–277 (2000).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Berardini, T. Z. et al. The Arabidopsis information resource: making and mining the ‘gold standard’ annotated reference plant genome. Genesis 53, 474–485 (2015).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Conesa, A. & Götz, S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832 (2008).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. & Kanehisa, M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007).
Zwaenepoel, A. & Van de Peer, Y. wgd—simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics 35, 2153–2155 (2019).
Enright, A. J. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Proost, S. et al. i-ADHoRe 3.0—fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 40, e11 (2012).
Lynch, M. & Conery, J. S. The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3, 35–44 (2003).
Bowman, J. L. et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304 (2017).
Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008).
Jo Ann Banks. et al. The compact Selaginella genome identifies changes in gene content associated with the evolution of vascular plants. Science 332, 960–963 (2011).
Liu, H. et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 7, 748–756 (2021).
Zimin, A. V. et al. An improved assembly of the loblolly pine mega-genome using long-read single-molecule sequencing. Gigascience 6, 1–4 (2017).
Nystedt, B. et al. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584 (2013).
Albert, V. A. et al. The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013).
Gonzali, S. et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Huang, S. et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281 (2009).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Yu, J. et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3, 0266–0281 (2005).
Liu, J. et al. Gapless assembly of maize chromosomes using long-read technologies. Genome Biol. 21, 121 (2020).
Cheng, S. et al. Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179, 1057–1067 (2019).
Hori, K. et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5, 3978 (2014).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Zheng, Y. et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670 (2016).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Shannon, P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Finn, R. D., Miller, B. L., Clements, J. & Bateman, A. iPfam: a database of protein family and domain interactions found in the Protein Data Bank. Nucleic Acids Res. 42, D364–D373 (2014).
Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496 (2018).
Kall, L., Krogh, A. & Sonnhammer, E. L. L. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35, W429–W432 (2007).
Lewis, T. E. et al. Gene3D: extensive prediction of globular domains in proteins. Nucleic Acids Res. 46, D435–D439 (2018).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Acknowledgements
We thank X. Zhang (Institute of Botany, Chinese Academy of Sciences) for providing helpful advice and suggestions, G. Rao (Peking University) for providing the homozygous A. capillus-veneris plant and DNA sequencing sample, Y. Wu (AGIS, CAAS) for providing the lncRNA determination pipeline, J. Zhao (Boyce Thompson Institute) for converting the references to a uniform format, and W. Wang and L. Xu (Tsinghua University) for assistance with phytohormone and metabolites detection. This work was supported by the National Key Research and Development Program of China (grant no. 2019YFA0906200 to S.H.); Science, Technology and Innovation Commission of Shenzhen Municipality (grant no. ZDSYS20200811142605017 to J.Y.); the Elite Young Scientists Program of CAAS (to J.Y.); the Agricultural Science and Technology Innovation Program (to J.Y.); the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant no. 833522 to Y.V.d.P.); Methusalem funding from Ghent University (grant no. BOF.MET.2021.0005.01 to Y.V.d.P.); a postdoctoral fellowship from the Special Research Fund of Ghent University (grant no. BOFPDO2018001701 to Z.L.); and the Research Foundation – Flanders (FWO) (grant no. 3G032219 to H.C.).
Author information
Authors and Affiliations
Contributions
S.H., S.B., Y.F. and J.Y. conceived the study. Y.F. and J.Y. designed and managed the major scientific objectives. S.H., Y.F., W.J.L. and J.Y. coordinated the project. Y.F., W.J., Y. Yuan, L.W., X.Y. and X.Q. managed the plant materials. Q.Z. and P.S. assembled the genome and estimated the genome size. Y.F., Q.L., X.L., X.Q., Q.Z., P.S., L.W. and Z.Z. annotated the genomes. H.C., Z.L. and Y.V.d.P performed the WGD calling. Y.F., Q.L. and X.L. identified the repetitive elements, non-coding RNAs and lncRNAs. Y.F., Q.L., Y. Yan, R.Z., J.Z. and S.C. clustered the gene families and conducted the related phylogenetic analysis. Y.F. and Q.L. constructed the co-expression network of AdcLEC1. Y.F., Q.L., X.Q. and X.L. carried out the RNA-seq analysis on homosporous development and jasmonate biosynthesis genes. J.Y. and R.D. performed the jasmonate biosynthesis and signalling analyses. R.D., Y.F., W.J. and Y. Yuan contributed to hormone and metabolome sample preparation. R.D. carried out metabolome detection and characterized the function of coronatine-inducible metabolites. Y.F. and J.Y. led the article preparation, together with S.H., W.J.L., Y.V.d.P., Q.L., R.D., Z.L., X.Q., X.L. and X.Z. All authors read and approved the final article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Yuannian Jiao, Fay-Wei Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analyses of whole genome duplication (WGD) events.
a, Self-alignment dot plot based on paralogous pairs in collinear blocks. b, The distribution of KS of the whole paranome in A. capillus-veneris. c, The distribution of KS of the syntenic paralogous pairs in A. capillus-veneris. d, The analysis of ksrates for the whole paranome of A. capillus-veneris before rate adjustment. Light grey histogram and kernel density estimation (KDE) curve are plotted for the whole paranome KS distribution. The estimated mean mode and standard deviation from 200 bootstrapped KDEs of each orthologous distribution between A. capillus-veneris and other species is shown as dashed red lines and boxes with corresponding KS values denoted in the legend. e, The analysis of ksrates for the whole paranome of A. capillus-veneris after rate adjustment. The whole paranome KS distributions are overlaid with rate-adjusted divergence events in coloured vertical lines and boxes. The overall mixture model in the dark solid KDE curve consists of an exponential component in dotted grey curve and optimized log-normal components in dashed grey curves. Each log-normal component is labelled with a letter, shown as vertical dashed grey lines with circular labels. Rate-adjusted mode estimates of orthologous KS distributions between A. capillus-veneris and other species, representing speciation events, are drawn as numbered vertical long-dashed lines and associated coloured boxes showing the mean and the standard deviation. Lines representing the same speciation event in the phylogeny share colour and numbering while here only one speciation event is involved with A. capillus-veneris. Horizontal arrows in figure legends ‘divergence with’ part indicate the KS shifts resulted from the substitution rate adjustments. f, The analysis of ksrates for anchor pairs of A. capillus-veneris after rate adjustment. The KS distributions of two anchor pair clusters, namely a and b in filled blue and red KDE curves with associated peak, derived from the lognormal mixture modeling of median KS values for the collinear segment pairs, are shown as a grey histogram. Rate-adjusted mode estimates of orthologous KS distributions between A. capillus-veneris and other species, representing speciation events, are drawn as numbered vertical long-dashed lines, and associated coloured boxes showing the mean and the standard deviation. Lines representing the same speciation event in the phylogeny share colour and numbering while here only one speciation event is involved with A. capillus-veneris. Horizontal arrows in figure legends ‘divergence with’ part indicate the KS shifts resulted from the substitution rate adjustments.
Extended Data Fig. 2 Comparison of the genome features of A. capillus-veneris with those of A. filiculoides and S. cucullata.
a, Lengths of different repeat components in the indicated eight representative species across land plants. b, Distributions of nucleotide distance (D) calculated for LTR among three ferns, A. capillus-veneris, A. filiculoides and S. cucullata. c, Violet plot showing gene characteristics of ferns. The upper and lower edge of white frame in the violin plot represent the 75% and 25% quartiles, the central line denotes the median value, and the black square shows the mean value. The upper and lower terminal of line in the violin indicated the upper adjacent value and lower adjacent value, 1.5 × the interquartile range and outliers (solid points).
Extended Data Fig. 3 Time course of the mechanical wound-induced response of JA in Ginkgo biloba.
Leaves of G. biloba were wounded with a hemostat. Damaged leaves were harvested at the indicated time points after wounding and analysed for jasmonates (the precursor OPDA [▲], jasmonic acid (JA) [●], and JA-Ile [■]) accumulation by HPLC-MS; each data point represents the mean ± SD of five biological replicates.
Extended Data Fig. 4 The maximum-likelihood tree of the BRI1-BRL and EMS1 gene family in land plants.
The domains of BRI1-BRL (TM, LRR, ID, and KD) and its closest gene family EMS1 (TM, LRR, and KD) were identified from all main land plant groups (in different colours). Maximum-likelihood tree was constructed with parameters: WAG + F + R6 model and 1,000 bootstrap replicates. Whole genome assemblies from 24 species were used for identification of BRI1-BRL and EMS1 homologs, including 9 bryophytes (Mpo, Marchantia polymorpha; Cpl, Calohypnum plumiforme; Fan, Fontinalis antipyretica; Cpu, Ceratodon purpureus; Ppa, Physcomitrella patens; Psc, Pleurozium schreberi; Aan, Anthoceros angustus; Aag, Anthoceros agrestis; Apu, Anthoceros punctatus), 3 lycophytes (Ita, Isoetes taiwanensis; Smo, Selaginella moellendorffii; Sle, Selaginella lepidophylla), 3 ferns (Adc, Adiantum capillus-veneris; Afi, Azolla filiculoides; Scu, Salvinia cucullata), and 9 seed plants (Pab, Picea abies; Pta, Pinus taeda; Gbi, Ginkgo biloba; Ath, Arabidopsis thaliana; Atr, Amborella trichopoda; Csa, Cucumis sativus; Osa, Oryza sativa; Vvi, Vitis vinifera; Zma, Zea mays). The detailed information is provided in Supplementary Data 19.
Extended Data Fig. 5 The island domain of the BRI1-BRL gene family in land plant.
The island domains were extracted from peptide of BRI1-BRL homologues and aligned by MUSCLE. The conserved sites were indicated with red boxes.
Supplementary information
Supplementary Information
Supplementary Text, Figs. 1–5 and Tables 1–14.
Supplementary Data 1
Supplementary Data 1–20.
Source data
Source Data Fig. 3
Statistical source data for Fig. 3b,c.
Source Data Fig. 4
Statistical source data for Fig. 4d,e.
Source Data Fig. 5
Statistical source data for Fig. 5.
Rights and permissions
About this article
Cite this article
Fang, Y., Qin, X., Liao, Q. et al. The genome of homosporous maidenhair fern sheds light on the euphyllophyte evolution and defences. Nat. Plants 8, 1024–1037 (2022). https://doi.org/10.1038/s41477-022-01222-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01222-x
This article is cited by
-
Expression divergence of expansin genes drive the heteroblasty in Ceratopteris chingii
BMC Biology (2023)
-
Rickettsial DNA and a trans-splicing rRNA group I intron in the unorthodox mitogenome of the fern Haplopteris ensiformis
Communications Biology (2023)
-
The first homosporous lycophyte genome revealed the association between the recent dynamic accumulation of LTR-RTs and genome size variation
Plant Molecular Biology (2023)