Abstract
Plant roots acquire nutrients and water while managing interactions with the soil microbiota. The root endodermis provides an extracellular diffusion barrier through a network of lignified cell walls called Casparian strips, supported by subsequent formation of suberin lamellae. Whereas lignification is thought to be irreversible, suberin lamellae display plasticity, which is crucial for root adaptative responses. Although suberin is a major plant polymer, fundamental aspects of its biosynthesis and turnover have remained obscure. Plants shape their root system via lateral root formation, an auxin-induced process requiring local breaking and re-sealing of endodermal lignin and suberin barriers. Here, we show that differentiated endodermal cells have a specific, auxin-mediated transcriptional response dominated by cell wall remodelling genes. We identified two sets of auxin-regulated GDSL lipases. One is required for suberin synthesis, while the other can drive suberin degradation. These enzymes have key roles in suberization, driving root suberin plasticity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data to support the conclusions of this manuscript are included in the main text and the supplementary materials. The full RNA-seq dataset was deposited in the NCBI Gene Expression Omnibus under accession GSE153478. Source data are provided with this paper.
References
Castrillo, G. et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017).
Duran, P. et al. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983 (2018).
Hassani, M. A., Duran, P. & Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 6, 58 (2018).
Banda, J. et al. Lateral root formation in Arabidopsis: a well-ordered LRexit. Trends Plant Sci. 24, 826–839 (2019).
Stoeckle, D., Thellmann, M. & Vermeer, J. E. Breakout-lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 41, 67–72 (2018).
Andersen, T. G. et al. Tissue-autonomous phenylpropanoid production is essential for establishment of root barriers. Curr. Biol. (in the press).
Barberon, M. et al. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164, 447–459 (2016).
Li, B. et al. Role of LOTR1 in nutrient transport through organization of spatial distribution of root endodermal barriers. Curr. Biol. 27, 758–765 (2017).
Yadav, V. et al. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 26, 3569–3588 (2014).
Vermeer, J. E. et al. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343, 178–183 (2014).
Tian, Q., Uhlir, N. J. & Reed, J. W. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14, 301–319 (2002).
Fukaki, H., Tameda, S., Masuda, H. & Tasaka, M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168 (2002).
Swarup, K. et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954 (2008).
Lewis, D. R. et al. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell 25, 3329–3346 (2013).
Voß, U. et al. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat. Commun. 6, 7641 (2015).
Bakan, B. & Marion, D. Assembly of the cutin polyester: from cells to extracellular cell walls. Plants 6, 57 (2017).
Girard, A. L. et al. Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24, 3119–3134 (2012).
Naseer, S. et al. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl Acad. Sci. USA 109, 10101–10106 (2012).
Philippe, G. et al. Ester cross-link profiling of the cutin polymer of wild-type and cutin synthase tomato mutants highlights different mechanisms of polymerization. Plant Physiol. 170, 807–820 (2016).
Yeats, T. H. et al. The identification of cutin synthase: formation of the plant polyester cutin. Nat. Chem. Biol. 8, 609–611 (2012).
Berhin, A. et al. The root cap cuticle: a cell wall structure for seedling establishment and lateral root formation. Cell 176, 1367–1378 (2019).
Philippe, G. et al. Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 55, 11–20 (2020).
Andersen, T. G. et al. Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature 555, 529–533 (2018).
Doblas, V. G. et al. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355, 280–284 (2017).
Fujita, S. et al. SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J. 39, e103894 (2020).
Lucas, M., Godin, C., Jay-Allemand, C. & Laplaze, L. Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J. Exp. Bot. 59, 55–66 (2008).
Péret, B. et al. Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 14, 991–998 (2012).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Gasperini, D. et al. Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genet. 11, e1005300 (2015).
Siligato, R. et al. MultiSite Gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 170, 627–641 (2016).
Fauser, F., Schiml, S. & Puchta, H. Both CRISPR–Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359 (2014).
Li-Beisson, Y. et al. Acyl-lipid metabolism. Arabidopsis Book 11, e0161 (2013).
Ursache, R., Andersen, T. G., Marhavy, P. & Geldner, N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 93, 399–412 (2018).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Jan, M., Gobet, N., Diessler, S., Franken, P. & Xenarios, I. A multi-omics digital research object for the genetics of sleep regulation. Sci. Data 6, 258 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Alexa, A. & Rahnenfuhrer, J. topGO: enrichment analysis for gene ontology. R package version 2.38.1 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank C. Grefen and M. Barberon for insightful discussions about GDSL-domain-containing proteins, experimental approaches and stimulating discussions; and the Electron Microscopy Facility and Imaging Facility of the University of Lausanne and the Center of Microcopy and Image Analysis of the University of Zurich for excellent service and support. Work in the Geldner laboratory was supported by an ERC Consolidator Grant (GA-N: 616228-ENDOFUN) and two consecutive SNSF grants (CRSII3_136278 and 31003A-156261). R.U. was supported by an EMBO Long-Term Fellowship (EMBO ALTF 1046‐2015). Work in the Nawrath laboratory was supported by a Swiss National Science Foundation grant (no. 310030_188672/1). Work in the Vermeer laboratory was supported by grants from the Swiss National Science Foundation (Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung; PP00P3_157524 and 316030_164086), the Netherlands Organization for Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek; NWO 864.13.008) and support from the University of Neuchâtel.
Author information
Authors and Affiliations
Contributions
R.U., N.G. and J.E.M.V. conceived, designed and coordinated the project. R.U., C.D.J.V.T., V.D.T., D.D.B., K.G., V.S., T.G.A. and J.E.M.V. performed all experimental work. E.S.-S., S.C. and S.P. analysed RNA-seq data. J.E.M.V. wrote the first draft of the manuscript. R.U., T.G.A., C.N., N.G. and J.E.M.V. revised the manuscript and all authors were involved in the discussion of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Owen Rowland and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SHY2pro::NLS-3xmVENUS dynamics in Col-0 (a), slr-1 (b) and CASP1pro::shy2-2/slr-1 treated with NAA.
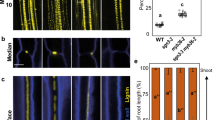
a. Maximum image projections of roots expressing SHY2pro::NLS-3xmVENUS in Col-0 (a), slr-1 (b) and CASP1pro::shy2-2/slr-1 (c) after 2, 4, 8, 16 and 24 hours of NAA treatment. Yellow dots indicate SHY2pro::NLS-3xmVENUS signal in the endodermis. The images in are representatives of each experiment repeated 3 times. Scale bar in (a) = 50 µm.
Extended Data Fig. 2 Differentiated endodermal cells have a distinct transcriptional response to auxin.
a, Confocal images of a root expressing SLRpro::CITRINE:SYP122 confirming that SLR is expressed in the epidermis, cortex, pericycle and weakly in the stele, but not in the endodermis (indicated by asterisks). The images are representatives of the experiment repeated 3 times. b and c, Comparison of the current dataset (Ursache) with the data set of Lewis et al.14. b, Heatmap showing that both datasets cluster separately and do not have significant overlap, which is confirmed by the analysis of correlation between the two data sets (c). d and e, Comparison of the current dataset (Ursache) with the data set of Voß et al.15. d, Heatmap showing that both datasets cluster separately and do not have significant overlap, which is confirmed by the analysis of correlation between the two data sets (e). p values in c and e are derived from a Pearson correlation test, are two-sided t-test values and not corrected for multiple testing. The corresponding cut-points (*/**/***) represent here p < 0.05, p < 0.01 and p < 0.001. The r value measures the correlation between 2 sets of data. The p-value states whether there are enough observations to believe that an observed correlation is not appearing by chance. Scale bar = 25 µm.
Extended Data Fig. 3 A high number of differentially regulated genes are expressed in the endodermis.
a, Confocal images of GELP12pro::NLS-3xmVENUS expression in different genetic backgrounds, showing repression in slr-1 under control conditions. b, Auxin treatment (10 µM NAA, 16hrs) results in induction of GELP12pro::NLS-3xmVENUS expression in the endodermis of slr-1 roots. NLS-3xmVENUS signal is shown in green and CFW staining of cell walls is shown in grey. c, Confocal images of roots expressing transcriptional markers of candidate genes differentially expressed between slr-1 and CASP1pro::shy2-2/slr-1 roots and showing specific expression in the endodermis. d, Confocal images showing xylem-specific expression of LAC2pro::NLS-3xmVENUS. e, Heatmap showing the expression dynamics of suberin-related genes significant differentially expressed. NLS-3xmVENUS signal is shown in green, CFW staining of cell walls in gray and cell wall staining by PI in red. The images in (a-d) are representatives of each experiment repeated at least 3 times. Scale bars = 20 µm.
Extended Data Fig. 4 Expression of suberin biosynthesis-related GELPs is induced by ABA and CIF2 treatment.
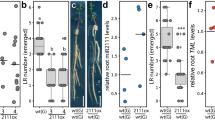
a, Confocal images showing effect of ABA treatment (1 µM, 24hr) on the expression domain of GPAT5pro::NLS-3xmVENUS and GELP38pro::NLS-3xmVENUS in Arabidopsis roots. The images are representatives of the experiment repeated 3 times. b, Quantification of the effect of ABA (1 µM, 24hr) and CIF2 peptide (500 nM, 24h) treatment on the expression of suberin biosynthesis-related GELPs identified as being repressed by auxin treatment. c, Schematic representation of the different single mutants of the suberin biosynthesis-related GELPs used in this study. d, qPCR results showing the relative gene expression in Col-0 control (100%) and T-DNA insertion lines of the suberin synthesis-related GELPs used in this study. The results are based on three biological replicates. The p value versus the Col-0 control for gelp49-1 is <0.000001 and for gelp51-1 is < 0.000001. ND, not detected. e, Quantification of suberin occupancy in the endodermis of the single mutants of the suberin biosynthesis-related GELPs using FY staining (n = 10 biologically independent samples). The p value versus the Col-0 control for gelp38-c1 is 0.1269, for gelp38-c2 is 0.2616, for gelp51-1 is > 0.9999, for gelp96-1 is 0.9385, for gelp22-1 is > 0.9999 and for gelp49-1 is > 0.9999. Different letters in (d) and (e) (p < 0.05) indicate statistically significant differences between means by ANOVA and two-sided t-test analysis. f, Schematic representation of the mutations in the gelpquint-1 and gelpquint-2 mutants. The mutations are indicated in red and the PAM sites in blue. Error bars in (b), (d) and (e) are SD. Scale bar in (a) = 25 µm.
Extended Data Fig. 5 Extended characterization of the phenotype of gelpquint mutants.
a, Nile Red staining of wild-type and gelpquint-1 and gelpquint-2 roots confirms the absence of suberin in the mutants. b, TEM micrographs of high-pressure frozen roots of wild-type and gelpquint-1 showing the absence of suberin lamella in the mutant. A region without lateral root primordia has been chosen to highlight the absence of suberin lamella. c, FY staining of CIF2 peptide treated Col-0 and gelpquint-1 seedlings showing absence of CIF2-mediated suberin deposition in gelpquint-1. d, TEM micrograph of high-pressure frozen roots of wild-type and gelpquint-1 showing that the lateral root cap cuticle is not affected in the gelpquint-1 mutant. e, Fluorescein di-acetate (FDA) uptake assay in wild-type roots showing a suberin mediated block of uptake at the level of the endodermis. f, Confocal image of a root expressing GELP38-XVEpro::GELP38- mCITRINE (green) in gelpquint-1 after β-Estradiol treatment (5 µM) stained with FM4-64 dye. g, TEM micrographs of roots of gelpquint-1 and the complementation by GELP38-XVEpro::GELP38- mCITRINE showing the complete recovery of endodermis suberin lamella. The images in (a-g) are representatives of each experiment repeated at least 3 times. h, Counting of PI-stained cells as a proxy for Casparian strip barrier in the roots of wild-type and gelpquint-1 and gelpquint-2 seedlings. All individual data points are plotted. No statistically significant difference was detected in using ANOVA and Bonferroni-adjusted paired two-sided t-test. The p value versus the Col-0 control for gelpquint-1 is 0.1680 and for gelpquint-2 is > 0.9999. i-j, Basic Fuchsin staining of the Casparian strip in early and differentiated endodermal cells of wild-type and gelpquint-1 and gelpquint-2 roots. k, Salt stress assay showing that gelpquint-1 and gelpquint-2 mutant seedlings are more sensitive to mild salt stress (85 mM NaCl) compared to wild-type. The images in (i-k) are representatives of each experiment repeated at least 3 times. l, Quantification of the effect of prolonged salt stress on the fresh weight of wild-type and gelpquint-1 and gelpquint-2 seedlings. All individual data points are plotted. The p value versus the Col-0 control for Col-0 transferred to salt is 0.7857, for gelpquint1 versus gelpquint1 transferred to salt is < 0.000001 and for gelpquint-2 versus gelpquint-2 transferred to salt is < 0.000001. m, Quantification of emerged lateral roots in wild-type and gelpquint-1 and gelpquint-2 mutants after 8 days of exposure to salt. All individual data points are plotted (n = 10). n, Quantification of the expression of known suberin biosynthesis-related genes in gelpquint-1 and gelpquint-2 mutants. Results are presented as fold-change compared to their expression levels in Col-0 (n = 9). No statistically significant difference was detected in using ANOVA and Bonferroni-adjusted paired two-sided t-test. For GPAT5, the p value versus Col-0 for gelpquint-1 is > 0.9999 and for gelpquint-2 is 0.4546, for ASFT the p values are > 0.9999 and > 0.9999; for HORST the p values are 0.1122 and > 0.9999; for FAR1 the p values are > 0.9999 and > 0.9999; for FAR4 the p values are 0.1467 and > 0.9999; for KSC2 the p values are 0.1136 and > 0.9999 correspondingly. Different letters in (m) (p < 0.001) and asterisks in (l) (p < 0.001) indicate statistically significant differences between means by ANOVA and two-sided t-test analysis. ns, not significant. For the boxplots in (l) and (m) the center depicts the median while the lower and upper box limits depict the 25th and 75th percentile respectively. Whiskers represent minima and maxima. Closed dots depict individual samples. Data in (h) and (n) are presented as mean +/- SD. Scale bars for (a), (d), (e), (f), (g), (h-j) = 25 µm. Scale bars for (b) and (c) = 1 µm, for (k) = 5 mm.
Extended Data Fig. 6 Auxin-upregulated GELPs show three distinct expression patterns.
a, FY staining of a Col-0 root at the site of lateral root emergence highlighting the presence of cuticle (indicated by arrow). b-c, FY staining of bodyguard mutant root at the site lateral root emergence. The absence of a proper cuticle is highlighted by the gap in FY staining. d-g, Confocal images showing the expression patterns of GELP12 (d), GELP72 (e), GELP73 (f) and GELP81 (g) during lateral root emergence. h-j, Confocal images showing the expression of GELP12 (h), GELP55 (i) and GELP72 (j) after 10 µM NAA treatment. NLS-3xmVENUS signal is in green and Calcofluor White staining of cell walls is in gray. The images are representatives of each experiment repeated at least 3 times. Scale bars = 25 µm.
Extended Data Fig. 7 Overexpression of three auxin-induced GELPs leads to suberin degradation.
a, FY staining on roots of Col-0 treated with b-Estradiol results in normal suberin pattern, whereas inducible endodermis-specific overexpression of GELP12, GELP55 or GELP72 results in degradation of suberin highlighted by absence of FY signal. The overexpression of GELP73 and GELP81 results in a normal suberin pattern similar to wild-type. b, FY staining of ELTPproXVE>>GELP12 lines germinated on normal 1/2MS medium for 4 days and transferred to the plates supplemented with 5 µM β-estradiol for 36 hours to observe the suberin degradation. c, TEM micrographs of ELTPproXVE>>GELP12 grown on plates with and without β-estradiol to highlight the absence of suberin lamella upon induction of GELP12 expression in endodermis. d, Schematic representation of the mutations in the auxin-upregulated single GELP mutants. The mutations are indicated in red and the PAM sites in blue. The images in (a-c) are representatives of each experiment repeated at least 3 times. Scale bars in (a) and (b) = 500 µm. Scale bars in (c) = 1 µm.
Extended Data Fig. 8 Some auxin-inducible GELPs facilitate lateral root emergence.
a-k, Gravistimulation-mediated induction of lateral root formation to functionally characterize the role of auxin-induced GELPs during lateral root formation. Staging of lateral root development was performed at 18hr and 42hr after gravistimulation. a, Col-0. b and c, gelp12-c1 and gelp12-c2. d and e, gelp55-c1 and gelp55-c2. f and g, gelp72-1 and gelp72-c1. h and i, gelp73-c1 and gelp73-c2. j and k, p values are indicated. A p value below 0.05 indicates a statistically significant difference as determined using a Pearson’s χ2 test. Experiments were repeated three times with a minimal of 15 seedlings per genotype and time point.
Supplementary information
Supplementary Information
Supplementary Tables 1–9.
Source data
Source Data Fig. 4
Statistical source data.
Rights and permissions
About this article
Cite this article
Ursache, R., De Jesus Vieira Teixeira, C., Dénervaud Tendon, V. et al. GDSL-domain proteins have key roles in suberin polymerization and degradation. Nat. Plants 7, 353–364 (2021). https://doi.org/10.1038/s41477-021-00862-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00862-9
This article is cited by
-
Ferric reduction by a CYBDOM protein counteracts increased iron availability in root meristems induced by phosphorus deficiency
Nature Communications (2024)
-
Choreographing root architecture and rhizosphere interactions through synthetic biology
Nature Communications (2024)
-
A suberized exodermis is required for tomato drought tolerance
Nature Plants (2024)
-
Identification, evolution, and expression of GDSL-type Esterase/Lipase (GELP) gene family in three cotton species: a bioinformatic analysis
BMC Genomics (2023)
-
Time course of changes in the transcriptome during russet induction in apple fruit
BMC Plant Biology (2023)