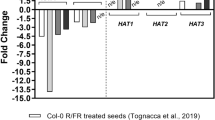
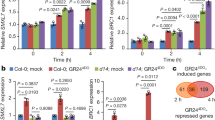
Abstract
Parasitic plant infestations dramatically reduce the yield of many major food crops of sub-Saharan Africa and pose a serious threat to food security on that continent1. The first committed step of a successful infestation is the germination of parasite seeds primarily in response to a group of related small-molecule hormones called strigolactones (SLs), which are emitted by host roots2. Despite the important role of SLs, it is not clear how host-derived SLs germinate parasitic plants. In contrast, gibberellins (GA) acts as the dominant hormone for stimulation of germination in non-parasitic plant species by inhibiting a set of DELLA repressors3. Here, we show that expression of SL receptors from the parasitic plant Striga hermonthica in the presence of SLs circumvents the GA requirement for germination of Arabidopsis thaliana seed. Striga receptors co-opt and enhance signalling through the HYPOSENSITIVE TO LIGHT/KARRIKIN INSENSITIVE 2 (AtHTL/KAI2) pathway, which normally plays a rudimentary role in Arabidopsis seed germination4,5. AtHTL/KAI2 negatively controls the SUPPRESSOR OF MAX2 1 (SMAX1) protein5, and loss of SMAX1 function allows germination in the presence of DELLA repressors. Our data suggest that ligand-dependent inactivation of SMAX1 in Striga and Arabidopsis can bypass GA-dependent germination in these species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data generated from this study are included in the article and Supplementary Tables. RNA-seq data from Fig. 3 were deposited in NCBI Sequence Read Archive under accession no. PRJNA602291, ID 602291.
References
Parker, C. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag. Sci. 65, 453–459 (2009).
Cook, C. E. et al. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190 (1966).
Sun, T. P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, R338–R345 (2011).
Waters, M. T. et al. Specialization within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 (2012).
Stanga, J. P., Smith, S. M., Briggs, W. R. & Nelson, D. C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163, 318–330 (2013).
Santner, A., Calderon-Villalobos, L. I. & Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307 (2009).
Lumba, S., Holbrook-Smith, D. & McCourt, P. The perception of strigolactones in vascular plants. Nat. Chem. Biol. 13, 599–606 (2017).
Waters, M. T., Gutjahr, C., Bennett, T. & Nelson, D. C. Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322 (2017).
Waldie, T., McCulloch, H. & Leyser, O. Strigolactones and the control of plant development: lessons from shoot branching. Plant J. 79, 607–622 (2014).
Cardoso, C., Ruyter-Spira, C. & Bouwmeester, H. J. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Sci. 180, 414–420 (2011).
Akiyama, K., Matsuzaki, K. & Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 (2005).
Yoshida, S. & Shirasu, K. Plants that attack plants: molecular elucidation of plant parasitism. Curr. Opin. Plant Biol. 15, 708–713 (2012).
Tsuchiya, Y. et al. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868 (2015).
Toh, S. et al. Structure–function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207 (2015).
Conn, C. et al. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543 (2015).
Scaffidi, A. et al. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165, 1221–1232 (2014).
Yao, R. et al. ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res. 27, 838–841 (2017).
Bennett, T. et al. Strigolactone regulates shoot development through a core signalling pathway. Biol. Open 5, 1806–1820 (2016).
Lantzouni, O., Klermund, C. & Schwechheimer, C. Largely additive effects of gibberellin and strigolactone on gene expression in Arabidopsis thaliana seedlings. Plant J. 92, 924–938 (2017).
Wallner, E. S., López-Salmerón, V. & Greb, T. Strigolactone versus gibberellin signaling: reemerging concepts? Planta 243, 1339–1350 (2016).
de Saint Germain, A. et al. Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol. 163, 1012–1025 (2013).
Tsuchiya, Y., Yoshimura, M. & Hagihara, S. The dynamics of strigolactone perception in Striga hermonthica: a working hypothesis. J. Exp. Bot. 69, 2281–2290 (2018).
Xie, X., Yoneyama, K. & Yoneyama, K. The strigolactone story. Annu. Rev. Phytopathol. 48, 93–117 (2010).
Lee, S. et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658 (2002).
Bassel, G. W. et al. Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl Acad. Sci. USA 108, 9709–9714 (2011).
Cao, D. et al. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142, 509–525 (2006).
Yang, Z. et al. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol. 32, 767–790 (2015).
Flematti, G. R., Ghisalberti, E. L., Dixon, K. W. & Trengove, R. D. A compound from smoke that promotes seed germination. Science 305, 977 (2004).
Nelson, D. C. et al. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 107, 7095–7100 (2010).
Nelson, D. C. et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 108, 8897–8902 (2011).
Toh, S. et al. Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem. Biol. 21, 988 (2014).
Xu, Y. et al. Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun. 9, 1 (2018).
Wallner, E.-S. et al. Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 27, 1241–1247 (2017).
Nelson, D. C. et al. Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873 (2009).
Marín-de la Rosa, N. et al. Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166, 1022–1032 (2014).
Chung, M. H., Chen, M. K. & Pan, S. M. Floral spray transformation can efficiently generate Arabidopsis. Transgenic Res. 9, 471–486 (2000).
Harrison, S. J. et al. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2, 19 (2006).
Gomez-Roldan, V. et al. Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008).
Suzuki, Y., Kawazu, T. & Koyama, H. RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37, 542–544 (2004).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
de Hoon, M. J. L., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004).
Saldanha, A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004).
Long, S. S. et al. A simple staining method for observation of germinated Striga seeds. Seed Sci. Res. 18, 125–129 (2008).
Graeber, K. et al. A guideline to family-wide comparative state-of-the-art quantitative RT–PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell 23, 2045–2063 (2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Winter, D. et al. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718 (2007).
Acknowledgements
We thank D. Nelson for the gift of smax1, smax1; smxl2 and max2; smax1 seeds, and T. Sun for the RGL2 clone. We acknowledge the contribution of P. McCourt for discussions and feedback on our manuscript. We acknowledge the Arabidopsis Biological Resource Center for obtaining ga1 mutants and the Parasitic Plant Genome Project for publicly available Striga transcriptome datasets. This work was supported by the Natural Sciences and Engineering Research Council of Canada in the form of a Discovery Grant (no. 06752), an Accelerator Supplement (no. 507992) and a Research Tools and Instruments grant awarded to S.L.
Author information
Authors and Affiliations
Contributions
S.L. contributed to all aspects of the research. M.B. designed and performed experiments, and analysed and interpreted data. S.T. contributed to the general conception of the project, and performed RT–qPCR in Arabidopsis and physiological experiments on Striga. C.W. constructed transgenic Arabidopsis lines and performed cell biology experiments using confocal microscopy. Z.X. performed RT–qPCR experiments and contributed to physiological assays. G.L. analysed RNA-seq data from Arabidopsis and identified homologues of Arabidopsis genes in S. hermonthica sequence datasets. C.S.P.M. contributed compounds required to conduct physiological experiments. G.P., K.E.N., P.S. and J.D.L. contributed to the generation of Arabidopsis transgenic lines and performed germination assays. J.D.S. provided S. hermonthica sequences and helped identify Striga homologues. S.L. and M.B. wrote the manuscript. S.L. directed and supervised all of the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Tom Bennett, Yuichiro Tsuchiya and Ruifeng Yao for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8.
Supplementary Table 1
List of statistical values for Figs. and Supplementary Figs.
Supplementary Table 2
Transcriptome data and lists of genes, strains and primers used in this study.
Rights and permissions
About this article
Cite this article
Bunsick, M., Toh, S., Wong, C. et al. SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat. Plants 6, 646–652 (2020). https://doi.org/10.1038/s41477-020-0653-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0653-z
This article is cited by
-
Insights from multi-omics integration into seed germination of Taxus chinensis var mairei
Communications Biology (2023)
-
Structural analysis of a hormone-bound Striga strigolactone receptor
Nature Plants (2023)
-
Probing strigolactone perception mechanisms with rationally designed small-molecule agonists stimulating germination of root parasitic weeds
Nature Communications (2022)
-
Resistance against broomrapes (Orobanche and Phelipanche spp.) in vegetables: a comprehensive view on classical and innovative breeding efforts
Euphytica (2022)
-
Mechanisms of pre-attachment Striga resistance in sorghum through genome-wide association studies
Molecular Genetics and Genomics (2022)