Abstract
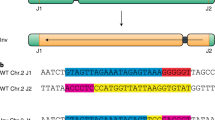
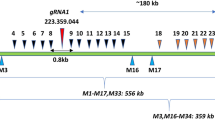
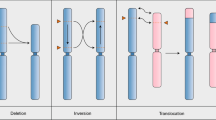
CRISPR–Cas is a powerful double-strand-break technology with wide-ranging applications from gene discovery to commercial product development. Thus far, this tool has been almost exclusively used for gene knockouts and deletions, with a few examples of gene edits and targeted gene insertions. Here, we demonstrate the application of CRISPR–Cas9 technology to mediate targeted 75.5-Mb pericentric inversion in chromosome 2 in one of the elite maize inbred lines from Corteva Agriscience. This inversion unlocks a large chromosomal region containing substantial genetic variance for recombination, thus providing opportunities for the development of new maize varieties with improved phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings are available within the paper or are available from the corresponding author upon reasonable request. Corteva Agriscience will provide plasmids to academic investigators for non-commercial research under an applicable material transfer agreement subject to proof of permission from any third-party owners of all or parts of the material and to governmental regulation considerations. Completion of a stewardship plan is also required. The Corteva Agriscience inbred line PH1V5T described in this research is proprietary.
References
Parisi, C., Tillie, P. & Rodríguez-Cerezo, E. The global pipeline of GM crops out to 2020. Nat. Biotechnol. 34, 31–36 (2016).
Raman, R. The impact of genetically modified (GM) crops in modern agriculture: a review. GM Crops Food 8, 195–208 (2017).
Voytas, D. F. Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350 (2013).
Gao, C. The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 19, 275–276 (2018).
Huang, T.-K. & Puchta, H. CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep. 38, 443–453 (2019).
Schmidt, C., Pacher, M. & Puchta, H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 98, 577–589 (2019).
Beying, N., Schmidt, C., Pacher, M., Houben, A. & Puchta, H. CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 6, 638–645 (2020).
Schmidt, C., Pacher, M. & Puchta, H. From gene editing to genome engineering: restructuring plant chromosomes via CRISPR/Cas. aBIOTECH 1, 21–31 (2020).
Lee, K. & Wang, K. Level up to chromosome restructuring. Nat. Plants 6, 600–601 (2020).
Puchta, H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14 (2005).
Lowry, D. B. & Willis, J. H. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8, e1000500 (2010).
Huang, K. & Rieseberg, L. H. Frequency, origins, and evolutionary role of chromosomal inversions in plants. Front. Plant Sci. https://doi.org/10.3389/fpls.2020.00296 (2020).
Brunner, S., Fengler, K., Morgante, M., Tingey, S. & Rafalski, A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17, 343–360 (2005).
Beló, A. et al. Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor. Appl. Genet. 120, 355–367 (2009).
Sun, S. et al. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 50, 1289–1295 (2018).
Liu, J. et al. Gapless assembly of maize chromosomes using long read technologies. Genome Biol. 21, 121 (2020).
Ou, S. et al. Effect of sequence depth and length in long-read assembly of the maize inbred NC358. Nat. Commun. 11, 2288 (2020).
Song, J.-M. et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45 (2020).
Liu, Y. et al. Pan-genome of wild and cultivated soybeans. Cell 182, 162–176 (2020).
Tao, Y., Zhao, X., Mace, E., Henry, R. & Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 12, 156–169 (2019).
Lowe, K. et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015 (2016).
Lowe, K. et al. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 54, 240–252 (2018).
Mak, A. A. Y. et al. Genome-wide structural variation detection by genome mapping on nanochannel arrays. Genetics 202, 351–362 (2016).
Svitashev, S. K., Pawlowski, W. P., Makarevitch, I., Plank, D. W. & Somers, D. A. Complex transgene locus structures implicate multiple mechanisms for plant transgene rearrangement. Plant J. 32, 433–445 (2002).
Svitashev, S. et al. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Phys. 169, 931–945 (2015).
Zhao, Y., Qian, Q., Wang, H. & Huang, D. Hereditary behavior of bar gene cassette is complex in rice mediated by particle bombardment. J. Genet. Genomics 34, 824–835 (2007).
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K. & Cigan, A. M. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274 (2016).
Jones, T. et al. Maize transformation using the morphogenic genes Baby boom and Wuschel2. Methods Mol. Biol. 1864, 81–93 (2019).
Acknowledgements
We thank G. St Clair, S. Jones and A. Ulm for assistance with the transformation experiments, M. Yang for help with the molecular analysis of T1 progeny, T. Engelhart for cultivation of the plants in the greenhouse, and M. Fedorova, J. Gerke and W. Gordon-Kamm for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
S.S. conceived the project, and C.S., K.F. and S.S designed the experiments. C.S., P.B., L.F. and B.L. conducted the experiments and V.L. performed the Bionano analysis. C.S., B.L., P.B. and S.S. analysed the data. K.F., V.L., P.B. and S.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors are employed by Corteva Agriscience.
Additional information
Peer review information Nature Plants thanks Kan Wang, Lanqin Xia and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Schwartz, C., Lenderts, B., Feigenbutz, L. et al. CRISPR–Cas9-mediated 75.5-Mb inversion in maize. Nat. Plants 6, 1427–1431 (2020). https://doi.org/10.1038/s41477-020-00817-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-00817-6
This article is cited by
-
Genome editing based trait improvement in crops: current perspective, challenges and opportunities
The Nucleus (2024)
-
Crop bioengineering via gene editing: reshaping the future of agriculture
Plant Cell Reports (2024)
-
Hidden prevalence of deletion-inversion bi-alleles in CRISPR-mediated deletions of tandemly arrayed genes in plants
Nature Communications (2023)
-
Genome-edited foods
Nature Reviews Bioengineering (2023)
-
Synthetic maize centromeres transmit chromosomes across generations
Nature Plants (2023)