Abstract
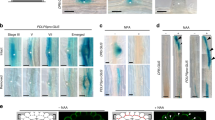
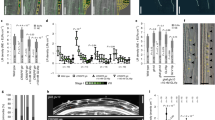
Lateral root (LR) emergence is a highly coordinated process involving precise cell–cell communication. Here, we show that MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3) and MPK6, and their upstream MAP-kinase kinases (MAPKKs), MKK4 and MKK5, function downstream of HAESA (HAE)/HAESA-LIKE2 (HSL2) and their ligand INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) during LR emergence. Loss of function of MKK4/MKK5 or MPK3/MPK6 results in restricted passage of the growing lateral root primordia (LRP) through the overlaying endodermal, cortical and epidermal cell layers, leading to reduced LR density. The MKK4/MKK5–MPK3/MPK6 module regulates the expression of cell wall remodelling genes in cells overlaying LRP and therefore controls pectin degradation in the middle lamella. Expression of constitutively active MKK4 or MKK5 driven by the HAE or HSL2 promoter fully rescues the LR emergence defect in the ida and hae hsl2 mutants. In addition, the MKK4/MKK5–MPK3/MPK6 module is indispensable in auxin-facilitated LR emergence. Our study provides insights into the auxin-governed and IDA–HAE/HLS2 ligand–receptor pair-mediated LR emergence process.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated or analysed in this study are included in this article and Supplementary Information files. The data that support the findings of this study are available from the corresponding author upon request.
References
Casimiro, I. et al. Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8(4), 165–171 (2003).
Nibau, C., Gibbs, D. J. & Coates, J. C. Branching out in new directions: the control of root architecture by lateral root formation. New Phytol. 179, 595–614 (2008).
Hochholdinger, F. & Zimmermann, R. Conserved and diverse mechanisms in root development. Curr. Opin. Plant Biol. 11, 70–74 (2008).
Petricka, J. J., Winter, C. M. & Benfey, P. N. Control of Arabidopsis root development. Plant Biol. 63, 563–590 (2012).
Malamy, J. E. & Benfey, P. N. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44 (1997).
Benková, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003).
Lavenus, J. et al. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458 (2013).
Vilchesbarro, A. & Maizel, A. Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 23, 31–38 (2015).
Peret, B. et al. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408 (2009).
Péret, B., Larrieu, A. & Bennett, M. J. Lateral root emergence: a difficult birth. J. Exp. Bot. 60, 3637–3643 (2009).
Lewis, D. R. et al. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell 25, 3329–3346 (2013).
Cai, S. & Lashbrook, C. C. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 146, 1305–1321 (2008).
Ogawa, M., Kay, P., Wilson, S. & Swain, S. M. Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233 (2009).
Swarup, K. et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954 (2008).
Kumpf, R. P. et al. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl Acad. Sci. USA 110, 5235–5240 (2013).
Cho, S. K. et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 105, 15629–15634 (2008).
Santiago, J. et al. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5, e15075 (2016).
Aalen, R. B., Wildhagen, M., Stø, I. M. & Butenko, M. A. IDA: a peptide ligand regulating cell separation processes in Arabidopsis. J. Exp. Bot. 64, 5253–5261 (2013).
Xu, J. & Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20, 56–64 (2015).
Cristina, M. S., Petersen, M. & Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649 (2010).
Tena, G., Boudsocq, M. & Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529 (2011).
Hamel, L.-P. et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198 (2006).
Meng, X. et al. Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 14, 1330–1338 (2016).
Xu, J. et al. A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234 (2014).
Xu, J. et al. Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 28, 1144–1162 (2016).
Su, J. et al. Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell 29, 526–542 (2017).
López-Bucio, J. S. et al. Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J. Exp. Bot. 65, 169–183 (2014).
Zhang, M. et al. Maternal control of embryogenesis by MPK6 and its upstream MKK4 and MKK5 in Arabidopsis. Plant J. 92, 1005–1019 (2017).
Müller, J. et al. Arabidopsis MPK6 is involved in cell division plane control during early root development, and localizes to the pre-prophase band, phragmoplast, trans-Golgi network and plasma membrane. Plant J. 61, 234–248 (2010).
Lucas, M. et al. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc. Natl Acad. Sci. USA 110, 5229–5234 (2013).
Micheli, F. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419 (2001).
Tian, G., Chen, M., Zaltsman, A. & Citovsky, V. Pollen-specific pectin methylesterase involved in pollen tube growth. Dev. Biol. 294, 83–91 (2006).
Schardon, K. et al. Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597 (2016).
Wang, H., Ngwenyama, N., Liu, Y., Walker, J. C. & Zhang, S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73 (2007).
Wang, H. et al. Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20, 602–613 (2008).
Guan, Y. et al. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 10, e1004384 (2014).
Guan, Y., Lu, J., Xu, J., McClure, B. & Zhang, S. Two mitogen-activated protein kinases, MPK3 and MPK6, are required for funicular guidance of pollen tubes in Arabidopsis. Plant Physiol. 165, 528–533 (2014).
Meng, X. et al. A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948–4960 (2012).
Peret, B. et al. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol. Syst. Biol. 9, 699–699 (2013).
Bush, S. M. & Krysan, P. J. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot. 58, 2181–2191 (2007).
Winter, D. et al. An electronic fluorescent pictograph browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718 (2007).
Meng, X. et al. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361–2372 (2015).
Bergmann, D. C., Lukowitz, W. & Somerville, C. R. Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497 (2004).
Liu, Y. & Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399 (2004).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Habets, M. E. J. & Offringa, R. PIN-driven polar auxin transport in plant developmental plasticity: a key target for environmental and endogenous signals. New Phytol. 203, 362–377 (2014).
Acknowledgements
This research was supported by the grants from Zhejiang Provincial Natural Science Foundation of China (no. LR18C020001), 111 project (no. B14027), National Natural Science Foundation of China (no. 31570297) and Zhejiang University Special Fund for Young Researchers (no. 2018QNA6002) to J.X. We thank W. Lukowitz (University of Georgia, Athens, Georgia, USA) for mkk4 and mkk5 single TILLING mutant seeds.
Author information
Authors and Affiliations
Contributions
Q.Z., J.X. and S.Z. designed the project. Q.Z., Y.S., S.G., M.Z., T.Z., X.H. and Y.L. ran the experiments. Q.Z., J.W., S.Z. and J.X. analysed the results. Q.Z., S.Z. and J.X. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information Nature Plants thanks Tom Beeckman, Melinka Butenko, Jean Colcombet and other anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–22 and Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Zhu, Q., Shao, Y., Ge, S. et al. A MAPK cascade downstream of IDA–HAE/HSL2 ligand–receptor pair in lateral root emergence. Nat. Plants 5, 414–423 (2019). https://doi.org/10.1038/s41477-019-0396-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0396-x
This article is cited by
-
Fossils and plant evolution: structural fingerprints and modularity in the evo-devo paradigm
EvoDevo (2022)
-
A peptide encoding gene MdCLE8 regulates lateral root development in apple
Plant Cell, Tissue and Organ Culture (PCTOC) (2022)
-
Receptor-like kinase HAESA-like 1 positively regulates seed longevity in Arabidopsis
Planta (2022)
-
Development of potent promoters that drive the efficient expression of genes in apple protoplasts
Horticulture Research (2021)
-
Genome-wide characterization and expression profiling of MAPK cascade genes in Salvia miltiorrhiza reveals the function of SmMAPK3 and SmMAPK1 in secondary metabolism
BMC Genomics (2020)