Abstract
Alternaria leaf spot in apple (Malus x domestica), caused by the fungal pathogen Alternaria alternata f. sp. mali (also called A. mali), is a devastating disease resulting in substantial economic losses. We previously established that the resistance (R) protein MdRNL2, containing a coiled-coil, nucleotide-binding, and leucine-rich repeat (CCR-NB-LRR) domain, interacts with another CCR-NB-LRR protein, MdRNL6, to form a MdRNL2–MdRNL6 complex that confers resistance to A. mali. Here, to investigate the function of the MdRNL2–MdRNL6 complex, we identified two novel pathogenesis-related (PR) proteins, MdPR10-1 and MdPR10-2, that interact with MdRNL2. Yeast two-hybrid (Y2H) assays and bimolecular fluorescence complementation (BiFC) assays confirmed that MdPR10-1 and MdPR10-2 interact with MdRNL2 and MdRNL6 at the leucine-rich repeat domain. Transient expression assays demonstrated that accumulation of MdPR10-1 and MdPR10-2 enhanced the resistance of apple to four strains of A. mali that we tested: ALT1, GBYB2, BXSB5, and BXSB7. In vitro antifungal activity assays demonstrated that both the proteins contribute to Alternaria leaf spot resistance by inhibiting fungal growth. Our data provide evidence for a novel regulatory mechanism in which MdRNL2 and MdRNL6 interact with MdPR10-1 and MdPR10-2 to inhibit fungal growth, thereby contributing to Alternaria leaf spot resistance in apple. The identification of these two novel PR proteins will facilitate breeding for fungal disease resistance in apple.
Similar content being viewed by others
Introduction
Apple Alternaria leaf spot caused by the fungal pathogen Alternaria alternata f. sp. mali (also called A. mali) is a devastating and economically major disease that causes severe early defoliation and weakens tree vigor, leading to reduced apple production1. In infected trees, brown or black leaf spots appear on the leaflets; these rapidly spread and fuse into brown or black necrotic lesions, causing leaves to fall off2,3. Fruits can also be infected, resulting in brown or black sunken lesions that do not increase in size over time2,3. Current management methods for Alternaria leaf spot primarily involve traditional chemical control agents, which are expensive and damaging to the environment. Identifying the molecular mechanisms involved in Alternaria leaf spot resistance in apple would set the stage for selecting for, or enhancing, these beneficial mechanisms4.
When fungi attack apple leaves, they typically produce specific toxins and effectors, which cause the accumulation of hydrogen peroxide (H2O2); the generation of reactive oxygen species and hormones, such as jasmonic acid, salicylic acid and ethylene, and the activation of defense-related genes, ultimately leading to cell death5,6. Plants have evolved a variety of disease resistance (R) proteins that mediate the recognition of pathogen effectors and activate downstream plant immunity signaling responses, enabling them to overcome fungal pathogen attack7. Fungal effectors that overcome PAMP-triggered immunity (PTI) are recognized by one of the R proteins predicted to encode intracellular proteins containing nucleotide-binding (NB) and leucine-rich repeat (LRR) domains, and the LRR domain is a conserved feature of many R proteins8. In pepper (Capsicum annuum), pathogenesis-related protein 10 (PR10) forms a complex with leucine-rich repeat protein 1 (LRR1), and silencing of PR10/LRR1 compromises resistance to avirulent Xanthomonas campestris pv vesicatoria infection; in contrast, overexpression of heterologous PR10 confers enhanced resistance in Arabidopsis9. Thus, the LRR1–PR10 complex is responsible for cell death-mediated defense signaling, and this role is strengthened by interaction with LRR19.
Pathogenesis-related (PR) genes are induced by pathogen infection, and their expression is associated with enhanced resistance to pathogens; thus, PR proteins may act as antimicrobial agents in defense signaling processes, such as cell wall hydrolysis and contact toxicity10. Based on their primary structure and biological activity, PR proteins have been grouped into 17 families10,11. According to sequence analysis, among the 17 families, the PR10 family is similar to a major birch (Betula alba) pollen allergen, Bet v 1, and has functions in antimicrobial activity, developmental processes, and secondary metabolism12,13,14,15,16,17. Several PR10 family genes have been identified from diverse plant species, including birch (Betula alba), asparagus (Asparagus officinalis), parsley (Petroselinum crispum), pea (Pisum sativum), common bean (Phaseolus vulgaris), potato (Solanum tuberosum), sorghum (Sorghum bicolor), rice (Oryza sativa), pepper, and apple15,17,18,19,20,21,22,23,24. Rice blast lesion mimic mutants Os-PR10a and Os-PR10b have high expression levels, and mutants show a spontaneous cell death phenotype25,26. Mal d 1 (PR10), an 18-kDa intracellular PR protein, is the major apple allergen in Central and Northern Europe. The expression of PR10 is induced by the microbial attack, fungal elicitors, and wounding stress in monocot asparagus, parsley, and bean22,23,24; however, the specific functions of PR10 in plant immunity signaling remain unclear10.
“Hanfu”, an apple cultivar commonly grown in China, is highly resistant to the A. mali strain ALT1. We previously established that HF plants inoculated with ALT1 accumulate MdRNL2 and are resistant to A. mali25. However, the role of MdRNL2 resistance in A. mali remains unclear. To further understand the MdRNL2 function, we performed semi–in vivo pull-down assays and liquid chromatography–mass spectrometry (LC–MS). In addition to proteins pulled down with an empty vector, there were 12 proteins in the LC–MS results, which were classified into three groups: plant defense, protein biosynthesis, and energy metabolism. From the LC–MS results, we established that the R protein MdRNL2 interacts with another R protein, MdRNL6, to form a complex through the NB-ARC domains of the proteins and that this interaction is necessary for resistance to A. mali (unpublished data). In addition to MdRNL6, we identified two PR10 members, which we named MdPR10-1 (major allergen Mal d 1-like; NCBI: XM_008352950.2) (Supplemental Figure 1A) and MdPR10-2 (major allergen Mal d 1; NCBI: NM_001294363.1) (Supplemental Figure 1B). To clarify the mechanism by which the MdRNL2–MdRNL6 complex inhibits ALT1 infection, we investigated the relationship among MdRNL2–MdRNL6, MdPR10-1, and MdPR10-2, as well as the roles of MdPR10-1 and MdPR10-2 in Alternaria leaf spot resistance. The identification of these two previously unknown proteins related to pathogenesis provides important insight that should facilitate breeding for resistance to fungal disease in apple.
Results
Identification of two novel pathogenesis-related proteins in apple: MdPR10-1 and MdPR10-2
Protein–protein interaction and LC–MS analyses identified two PR proteins, MdPR10-1 and MdPR10-2, that interact with MdRNL2 (Supplemental Figure 1). We cloned the cDNA sequences corresponding to MdPR10-1 and MdPR10-2 from HF and searched for them in the NCBI BLASTn database (https://blast.ncbi.nlm.nih.gov/). MdPR10-1 is located on chromosome 13, and MdPR10-2 is located on chromosome 16 (Supplemental Figure 2). Our phylogenetic analysis showed that MdPR10-1 and MdPR10-2 are closely related to PR10 gene sequences from other species. MdPR10-1 and MdPR10-2 are most closely related to PR10 gene sequences from pear (Pyrus bretschneideri), peach (Prunus persica), and sweet cherry (Prunus avium), revealing a high degree of conservation in these species. In contrast, pathogenesis-related protein 1 from Malus × domestica (MdPR1), which was used as an outgroup gene for phylogenetic analysis, was not closely related to these genes (Supplemental Figure 3). After deducing the protein sequences of MdPR10-1 and MdPR10-2 from the nucleotide sequences, we found that MdPR10-1 and MdPR10-2 are each 159 amino acids long (Supplemental Figure 4A) and share the GXGGXGXXK consensus sequence and the Bet v 1 motif, as determined by BLAST analysis (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SnapGene software using the PR10 signature as a query (Supplemental Figure 4A, B).
MdPR10-1 and MdPR10-2 physically interact with MdRNL2–MdRNL6
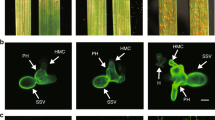
MdPR10-1 and MdPR10-2 were highly induced in HF by ALT1 inoculation, whereas the outgroup gene MdPR1 was not induced (Supplemental Figure 5). Next, we tested for interactions between MdRNL2–MdRNL6 and the two identified PR proteins using yeast two-hybrid (Y2H) assays. We cloned the MdPR10-1 and MdPR10-2 coding sequences into pGADT7 and pGBKT7 (yielding MdPR10-1-AD, MdPR10-2-AD, MdPR10-1-BK, and MdPR10-2-BK) and cotransfected the combinations MdRNL2-AD/MdPR10-1-BK, MdRNL2-AD/MdPR10-2-BK, MdPR10-1-AD/MdRNL6-BK, and MdPR10-2-AD/MdRNL6-BK into the yeast strain AH109 (Fig. 1A). Colonies containing any of the four combinations grew on synthetic dropout nutrient medium and were stained with X-α-gal, suggesting that MdPR10-1 and MdPR10-2 each interact with both MdRNL2 and MdRNL6 (Figs. 1A, B).
A Yeast two-hybrid (Y2H) analysis showing that both MdPR10-1 and MdPR10-2 interact with MdRNL2 and MdRNL6. Coexpression of SV40-AD/BD-P53 was used as a positive control. As negative controls, MdPR10-1-AD/BK, MdPR10-2-AD/BK, AD/MdPR10-1-BK, AD/MdPR10-2-BK, and AD/BD were coexpressed. B β-Galactosidase activity in Y2H assays. The error bars show the standard error of the mean (SEM) values (n = 3 biological replicates). **P < 0.01 (Student’s t test). C BiFC assay of the interactions between MdPR10-1 and MdRNL2–MdRNL6 and between MdPR10-2 and MdRNL2–MdRNL6. MdRNL2-YFPc/MdPR10-1-YFPn, MdRNL2-YFPc/MdPR10-2-YFPn, MdPR10-1-YFPc/MdRNL6-YFPn, and MdPR10-2-YFPc/MdRNL6-YFPn were transiently expressed in N. benthamiana epidermal cells. 1300-YFPc/MdPR10-1-YFPn, 1300-YFPc/MdPR10-2-YFPn, MdPR10-1-YFPc/1300-YFPn, and MdPR10-2-YFPc/1300-YFPn were used as negative controls. Scale bars = 50 μm
To verify the interaction of MdPR10-1 and MdPR10-2 with MdRNL2–MdRNL6 in plant cells, we conducted a bimolecular fluorescence complementation (BiFC) assay in Nicotiana benthamiana. The combinations MdPR10-1-YFPc/1300-YFPn, MdPR10-2-YFPc/1300-YFPn, MdPR10-1-YFPc/MdRNL6-YFPn, MdPR10-2-YFPc/MdRNL6-YFPn, 1300-YFPc/MdPR10-1-YFPn, 1300-YFPc/MdPR10-2-YFPn, MdRNL2-YFPc/MdPR10-1-YFPn, and MdRNL2-YFPc/MdPR10-2-YFPn were each transiently expressed in N. benthamiana leaves using Agrobacterium tumefaciens transfection (Fig. 1C). As expected, epidermal cells expressing MdPR10-1-YFPc/MdRNL6-YFPn, MdPR10-2-YFPc/MdRNL6-YFPn, MdRNL2-YFPc/MdPR10-1-YFPn, and MdRNL2-YFPc/MdPR10-2-YFPn showed strong fluorescence signals, demonstrating that MdPR10-1 and MdPR10-2 interact with both MdRNL2 and MdRNL6 in vivo (Fig. 1C). These results suggest that MdPR10-1 and MdPR10-2 physically bind to MdRNL2–MdRNL6 in apple.
MdPR10-1 and MdPR10-2 physically bind to MdRNL2–MdRNL6 at the LRR domain
To investigate whether the interactions of MdPR10-1 and MdPR10-2 with MdRNL2 and MdRNL6 involve direct physical binding and to identify the protein domains involved, we analyzed the pairwise interactions between various truncated versions of MdRNL2–MdRNL2-1 (CCR domain of MdRNL2), MdRNL2-2 (NB-ARC domain of MdRNL2), MdRNL2-3 (LRR domain of MdRNL2), MdRNL6-1 (CCR domain of MdRNL6), MdRNL6-2 (NB-ARC domain of MdRNL6) or MdRNL6-3 (LRR domain of MdRNL6) and MdPR10-1 or MdPR10-2 using the Y2H system. Coexpression of MdPR10-1-AD/MdRNL6-1-BK, MdPR10-1-AD/MdRNL6-2-BK, MdPR10-1-AD/MdRNL6-3-BK, MdPR10-2-AD/MdRNL6-1-BK, MdPR10-2-AD/MdRNL6-2-BK, MdPR10-2-AD/MdRNL6-3-BK, MdRNL2-1-AD/MdPR10-1-BK, MdRNL2-2-AD/MdPR10-1-BK, MdRNL2-3-AD/MdPR10-1-BK, MdRNL2-1-AD/MdPR10-2-BK, MdRNL2-2-AD/MdPR10-2-BK, and MdRNL2-3-AD/MdPR10-2-BK allowed growth on the selective media (Fig. 2A), and staining with X-α-gal revealed that MdPR10-1 and MdPR10-2 interacted with MdRNL2-3 and MdRNL6-3. These data indicate that the LRR domains of MdRNL2 and of MdRNL6 interact with MdPR10-1 and MdPR10-2 (Figs. 2A, B). We validated this finding through a BiFC assay. The combinations MdPR10-1-YFPc/MdRNL6-3-YFPn, MdPR10-2-YFPc/MdRNL6-3-YFPn, MdRNL2-3-YFPc/MdPR10-1-YFPn, and MdRNL2-3-YFPc/MdPR10-2-YFPn resulted in strong fluorescence signals in the N. benthamiana epidermal cell cytoplasm (Fig. 2C). Thus, MdPR10-1 and MdPR10-2 appear to bind to the LRR domains of MdRNL2 and MdRNL6.
A Y2H analysis showing the interactions between MdPR10-1 and MdRNL2-3, MdPR10-2 and MdRNL2-3, MdPR10-1 and MdRNL6-3, and MdPR10-2 and MdRNL6-3. B β-Galactosidase activity in Y2H assays. The error bars show the standard error of the mean (SEM) values (n = 3 biological replicates). **P < 0.01 (Student’s t test). C BiFC assay of the interactions between MdPR10-1 and MdRNL2-3, MdPR10-2 and MdRNL2-3, MdPR10-1 and MdRNL6-3, and MdPR10-2 and MdRNL6-3 when transiently expressed in Nicotiana benthamiana epidermal cells. Scale bars = 50 μm
MdPR10-1 and MdPR10-2 are both required for ALT1 resistance
We next investigated the functional relationship between the MdPR10-1 and MdPR10-2 during ALT1 infection in apple. We constructed a binary vector (pFGC5941) containing partial sequences of MdPR10-1 and MdPR10-2 and their corresponding reverse complementary sequences to form hairpin structures (RNAi-MdPR10-1 and RNAi-MdPR10-2). This produced small interfering RNAs that specifically silenced MdPR10-1 and MdPR10-2 when transiently expressed in HF via agroinfiltration (Fig. 3A). Four days after transformation, the expression levels of MdPR10-1 and MdPR10-2 were downregulated in RNAi-MdPR10-1 and RNAi-MdPR10-2 HF seedlings (Fig. 3B). Forty-eight hours postinoculation (hpi) with ALT1, the average percentage of leaf area affected by lesions was significantly higher in RNAi-MdPR10-1 and RNAi-MdPR10-2 HF seedlings than in wild-type controls (WT; uninfiltrated HF seedlings) or empty vector controls (EV; HF seedlings agroinfiltrated with empty vector), demonstrating that resistance to ALT1 infection was suppressed (Figs. 3C, D). Reverse transcription-quantitative PCR (RT-qPCR) analysis showed that the A. mali biomass was increased in the RNAi-MdPR10-1 and RNAi-MdPR10-2 HF lines, explaining their loss of resistance (Fig. 3E). The gene-silencing screen indicated that the MdRNL2–MdRNL6 complex is required for MdPR10-1- and MdPR10-2-mediated resistance to ALT1.
A Schematic of the constructs used for silencing of MdPR10-1 (RNAi-MdPR10-1) and MdPR10-2 (RNAi-MdPR10-2). B–D MdPR10-1 and MdPR10-2 transcript contents revealed by RT-qPCR B, infection symptoms C, relative lesion areas D, and relative biomass of A. mail E in the WT and empty vector (EV)-, RNAi-MdPR10-1-, and RNAi-MdPR10-2-infiltrated Hanfu (HF) plants at 48 hpi with ALT1. F Schematic of constructs used for overexpression of MdPR10-1 (OE-MdPR10-1) and MdPR10-2 (OE-MdPR10-2). G–J MdPR10-1 and MdPR10-2 contents revealed by RT-qPCR G, infection symptoms H, relative lesion areas I, and relative biomass of A. mail J in the WT and in EV-, OE-MdPR10-1-, and OE-MdPR10-2-infiltrated NGR196 plants at 48 hpi. The spore inoculum concentration was 2 × 105 CFU/mL. Error bars in B, D, E, G, I, and J = SDs; **P < 0.01 (Student’s t test). Similar results were obtained in three independent biological replicates
In contrast, transformation of the full-length MdPR10-1 or MdPR10-2 transcript into the susceptible apple cultivar NGR196 via A. tumefaciens infiltration (yielding OE-MdPR10-1 or OE-MdPR10-2 NGR196 seedlings, respectively) (Fig. 3F) resulted in increased expression of MdPR10-1 and MdPR10-2 4 d after infiltration (Fig. 3G). At 48 hpi with ALT1, the average percentage of leaf area affected by lesions was lower in OE-MdPR10-1 and OE-MdPR10-2 NGR196 plants than in control WT and EV-infiltrated NGR196 plants, indicating that their resistance to Alternaria leaf spot was enhanced (Figs. 3H, I). RT-qPCR analysis showed that AMT1 expression was decreased in OE-MdPR10-1 and OE-MdPR10-2 NGR196 lines (Fig. 3J), possibly explaining why they developed resistance. These data demonstrate that the accumulation of MdPR10-1 and MdPR10-2 enhances ALT1 resistance in the susceptible NGR196 cultivar.
MdPR10-1 and MdPR10-2 inhibit ALT1 growth in vitro
Next, we clarified the mechanism by which MdPR10-1 and MdPR10-2 contribute to ALT1 resistance and tested whether MdPR10-1 and MdPR10-2 have a direct effect on fungal growth. We purified the recombinant MdPR10-1 and MdPR10-2 proteins with glutathione S-transferase (GST) resin and detected them on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (Supplemental Figure 6). An in vitro antifungal activity assay using these purified proteins confirmed that ALT1 growth was strongly inhibited in the groups treated with MdPR10-1 or MdPR10-2 compared with the untreated control group (Fig. 4A). A quantitative analysis of the antifungal activity of MdPR10-1 and MdPR10-2 indicated that the area of ALT1 mycelium growth in potato dextrose agar (PDA) differed greatly between cultures treated with MdPR10-1 and MdPR10-2 and the negative controls (Fig. 4B), suggesting that MdPR10-1 and MdPR10-2 contribute to ALT1 resistance by inhibiting fungal growth.
A Growth inhibition of ALT1 treated with MdPR10-1 and MdPR10-2 proteins. Left: 20 μL of ALT1 sporangial suspension + 60 μL of 500 μg/μL purified MdPR10-1/MdPR10-2 proteins. Right: 20 μL of ALT1 sporangial suspension + 60 μL of 500 μg/μL of purified GST proteins. B Area of ALT1 mycelium growth in PDA, as determined using ImageJ. The spore suspension concentration was 2 × 105 CFU/mL. The data were collected on day 6 in PDA. Scale bars = 1 cm. Student’s t test: **P < 0.01
MdPR10-1 and MdPR10-2 are both required for GBYB2, BXSB5, and BXSB7 resistance
To further explore the contributions of MdPR10-1 and MdPR10-2 to Alternaria leaf spot resistance, we studied three other strains of A. mali: GBYB2, BXSB5, and BXSB7. We transformed the full-length transcripts of MdPR10-1 and MdPR10-2 into the susceptible apple cultivar NGR196 via A. tumefaciens infiltration (Fig. 3F). Four days after infiltration, we observed increased MdPR10-1 and MdPR10-2 expression levels in OE-MdPR10-1 and OE-MdPR10-2 NGR196 seedlings, respectively (Supplemental Figure 7). At 48 hpi with GBYB2, BXSB5, or BXSB7, the average percentage of leaf area affected by lesions was lower in OE-MdPR10-1 and OE-MdPR10-2 plants than in control WT or EV-infiltrated plants, indicating that their resistance to Alternaria leaf spot was enhanced (Fig. 5). RT-qPCR analysis showed that AMT1 expression was decreased in OE-MdPR10-1 and OE-MdPR10-2 NGR196 lines inoculated with GBYB2, BXSB5, or BXSB7 (Fig. 5), possibly explaining their development of resistance. These data demonstrate that the accumulation of MdPR10-1 and MdPR10-2 enhances resistance to GBYB2, BXSB5, and BXSB7 in the NGR196 cultivar.
A Infection symptoms of WT and EV-, OE-MdPR10-1-, and OE-MdPR10-2-infiltrated NGR196 plants at 48 hpi with GBYB2. B, C Relative lesion areas B and relative biomass of A. mail C in the leaves of these plants. D–F Infection symptoms D, relative lesion areas E, and relative biomass of A. mail F in the leaves of WT and EV-, OE-MdPR10-1-, and OE-MdPR10-2-infiltrated NGR196 plants at 48 hpi with BXSB5. G–I Infection symptoms G, relative lesion areas H, and relative biomass of A. mail (I) in WT and EV-, OE-MdPR10-1-, and OE-MdPR10-2-infiltrated NGR196 plants at 48 hpi with BXSB7 plants. Throughout, the spore inoculum concentrations were 2 × 105 CFU/mL. Error bars = SDs; **P < 0.01 (Student’s t test). Similar results were obtained from three independent biological replicates
MdPR10-1 and MdPR10-2 inhibit GBYB2, BXSB5, and BXSB7 growth in vitro
In vitro antifungal activity assays confirmed that GBYB2, BXSB5, and BXSB7 growth was strongly inhibited in the groups treated with the recombinant proteins MdPR10-1 and MdPR10-2 compared with the negative control group (Figs. 6A, C, and E). In a quantitative analysis of antifungal activity, the area of mycelium growth in PDA differed greatly in samples treated with purified MdPR10-1 or MdPR10-2 compared with samples treated with the negative control (Figs. 6B, D, and F). These observations suggest that MdPR10-1 and MdPR10-2 contribute to GBYB2, BXSB5, and BXSB7 resistance by inhibiting fungal growth.
A Growth inhibition of GBYB2 treated with the MdPR10-1 and MdPR10-2 proteins. Left: 20 μL of GBYB2 sporangial suspension + 60 μL of 500 μg/μL of purified MdPR10-1/MdPR10-2 proteins. Right: 20 μL of GBYB2 sporangial suspension + 60 μL of 500 μg/μL of purified GST proteins. B Area of GBYB2 mycelium growth in PDA, as determined using ImageJ. C Growth inhibition of BXSB5 treated with the MdPR10-1 and MdPR10-2 proteins. Left: 20 μL of BXSB5 sporangial suspension + 60 μL of 500 μg/μL purified MdPR10-1/MdPR10-2 proteins. Right: 20 μL of BXSB5 sporangial suspension + 60 μL of 500 μg/μL purified GST proteins. D Area of BXSB5 mycelium growth in PDA, as determined with ImageJ. E Growth inhibition of BXSB7 treated with the MdPR10-1 and MdPR10-2 proteins. Left: 20 μL of BXSB7 sporangial suspension + 60 μL of 500 μg/μL purified MdPR10-1/MdPR10-2 proteins. Right: 20 μL of BXSB7 sporangial suspension + 60 μL of 500 μg/μL purified GST proteins. F Area of BXSB7 mycelium growth in PDA, as determined with ImageJ. Throughout, the spore suspension concentration was 2 × 105 CFU/mL. The data were collected on day 6 in PDA. Scale bars = 1 cm. Student’s t-test: **P < 0.01
Discussion
Several PR-10 genes have been isolated in various species with distinct expression patterns in response to stress conditions26,27. The first reported PR10 gene from parsley established the PR-10 class of PR proteins22,28. Shortly afterward, according to their sequence homology with PR-10 proteins, other proteins found in birch17, celery29, apple30, and other fruits and vegetables were also included in the PR-10 class, most of which were induced by pathogens31. In this study, we demonstrate that apple pathogenesis-related protein 10 is pivotal for apple defense and responses against fungal attack. In apple, the MdPR10-1 and MdPR10-2 genes have been identified, and the expression of MdPR10-1 and MdPR10-2 is induced by A. mali4. Although PR10 is known to have ribonuclease and antimicrobial activity in plants, little is known about the specific functions of PR10 in apple13,15. In our study, transient expression assays demonstrated that accumulation of MdPR10-1 and MdPR10-2 enhanced resistance to ALT1, GBYB2, BXSB5, and BXSB7 in apple (Fig. 3 and Fig. 5). Using in vitro antifungal activity assays, we established that MdPR10-1 and MdPR10-2 contributed to Alternaria leaf spot resistance by inhibiting fungal growth (Fig. 4 and Fig. 6).
In N. benthamiana (tobacco), overexpression of PR10 alone does not induce a cell death response in leaves32,33. Additionally, the PR10 family is well characterized at the structural level34. Until now, there has been no relative experimental evidence of any physical interaction between PR10 and other proteins in apple. The Bet v 1 fold domain in PR10 is responsible for its interaction with several types of ligands, such as cytokinins, brassinosteroids, and flavonoids, and might also be involved in its binding to LRR1; for example, CaPR10 forms a complex with LRR1 in pepper, which is resistant to bacterial infection9,35,36,37,38. In our study, Y2H and BiFC assays confirmed that both the MdPR10-1 and MdPR10-2 interact with MdRNL2–MdRNL6 at the LRR domain (Fig. 1 and Fig. 2). We established that the LRR domains of MdRNL2–MdRNL6 physically interact with MdPR10-1 and MdPR10-2, enhancing PR10-triggered defense responses in apple (Fig. 1 and Fig. 2).
Alternaria leaf blotch caused by Alternaria spp. leads to significant losses in apple production. When A. mali attacks apple leaves, the leaves first develop brown or black blotches surrounded by a dark margin; then, the leaves turn yellow and abscise prematurely from the canopy39. Leaf blotch mostly affects high-value and very popular cultivars, such as the ‘Fuji’, ‘Royal Gala’, ‘Red Delicious’, and ‘Pink Lady’ cultivars39. A. mali, referred to as the apple Alternaria leaf blotch pathotype, has been reported in Japan40, China41, Korea42, the United States43, Russia44, Yugoslavia45, Iran46, Turkey47, and Brazil48. Few studies have investigated Alternaria leaf blotch control and resistance genes. Our data provide detailed evidence for a novel regulatory mechanism in which MdRNL2–MdRNL6 interact with MdPR10-1 and MdPR10-2 to inhibit fungal growth, thereby contributing to Alternaria leaf spot resistance in apple (Supplemental Figure 8). These data warrant an investigation into the role of PR10 in defense responses in apple and will facilitate breeding for fungal disease resistance in this species.
Materials and methods
Plant materials
Tissue culture seedlings of different apples (Malus domestica cv. ‘Hanfu’, HF; Malus domestica cv. NGR196) were planted on Murashige and Skoog (MS) medium containing 0.6 mg/L 6-benzylaminopurine (6-BA) and 0.15 mg/L 1-naphthylacetic acid (NAA). The temperature of the culture room was controlled at 24 ± 1 °C, and the photoperiod was set as 16 h/8 h light/dark under fluorescent lamps (Philips TL5 28 W/865). Every four weeks, the seedlings were transferred to a fresh medium, and the plants were used for fungal infection and Agrobacterium tumefaciens infiltration experiments.
Fungal infection assay
Four strains of A. mali, ALT1, GBYB2, BXSB5, and BXSB7, were cultured on potato dextrose agar (PDA) medium in the dark, and the temperature was controlled at 25 °C. After 6 days of growth, the spores were diluted in double-distilled water, and the final concentration was quantified as 2 × 105 CFU/mL via observation under a microscope (Olympus CX31). Usually, 4 d after A. tumefaciens infiltration, spore suspensions of ALT1, GBYB2, BXSB5, or BXSB7 pathogens were inoculated on 4-week-old apple leaves4.
The lesion size was measured as described previously49. First, we removed leaves from more than 10 inoculated A. mali apple plantlets (approximately 30–40 leaves) at 48 hpi, and images of the leaves were taken with a scale bar. The lesion sizes were then calculated using ImageJ software. The results for three independent biological replicates were obtained.
Calculation of the relative DNA content of ALT1/GBYB2/BXSB5/BXSB7 in apple was performed as described previously49,50. After 48 h of inoculation with A. mali, the infected apple leaves were collected, and four leaves from different infected seedlings were regarded as one sample. DNA of apple leaves was extracted and purified by using a DNeasy Plant Mini Kit (Qiagen, 69104). SuperReal PreMix Plus (SYBR Green) (Tiangen, FP205) was used for the following real-time PCR assay, and the relative DNA abundance was calculated by the 2−ΔΔCT method51. The reference gene was MdActin (NCBI XM_008365636.2). The specific primers for AMT1 and the reference gene are listed in Supplemental Table 2.
Protein purification
MdPR10-1 and MdPR10-2 were inserted into the pGEX-4T-1 vector and transformed into Transetta(DE3) chemically competent Escherichia coli cells, whose growth was then induced at 16 °C. After 16 hours, the cells were suspended in suspension buffer (10 mM Tris-HCl, pH 7.4, 30 mM NaCl). Samples of the E. coli suspension culture were pulverized with ultrasound for 20 minutes and then pelleted by centrifugation (12,000 g, 1 h, 4 °C). The collected supernatant was purified with GST resin (Life Technologies, G2879) at 4 °C for 1 hour. The supernatant was then discarded, and the remaining beads were rinsed with glutathione buffer three times before being used for further experiments.
Yeast two-hybrid (Y2H) assays
For Y2H assays, the coding sequences of MdRNL2, MdRNL2-1, MdRNL2-2, MdRNL2-3, MdPR10-1, and MdPR10-2 were fused with the pGADT7 vector (Clontech). The coding sequences of MdRNL6, MdRNL6-1, MdRNL6-2, MdRNL6-3, MdPR10-1, and MdPR10-2 were fused with the pGBKT7 vector (Clontech). Different combinations of pGBKT7 and pGADT7 vectors were cotransformed into the AH109 strain, and then the transformed strains were grown on SD/–Leu–Trp medium at 30 °C for 4–5 days. For each combination, three independent experiments were carried out. The primers are shown in Supplemental Table 1.
Bimolecular fluorescence (BiFC) assays
The coding sequences of MdRNL2, MdRNL2-1, MdRNL2-2, MdRNL2-3, MdRNL6, MdRNL6-1, MdRNL6-2, MdRNL6-3, MdPR10-1, and MdPR10-2 were amplified and fused with the coding sequence of the N-terminus of YFP (YFPn) or the C-terminus of YFP (YFPc) and inserted into vectors. Various combinations of YFPn and YFPc were transiently expressed in apple leaves by Agrobacterium tumefaciens infiltration52. Four days later, YFP fluorescence in leaves was observed and imaged at wavelengths of 500–542 nm using a laser scanning confocal microscope (Olympus BX61). The cloning primers are listed in Supplemental Table 2.
Agrobacterium tumefaciens infiltration
In overexpression experiments, full-length MdPR10-1 and MdPR10-2 sequences were cloned into the vector pFGC5941 (GenBank AY310901) with NcoI/BamHI restriction sites. The empty pFGC5941 vector was used as the control. Loss-of-function MdPR10-1 and MdPR10-2 constructs were produced by cloning their specific sequences and their partially specific reverse sequences into pFGC5941 using the NcoI/SawI and XbaI/BamHI restriction sites for MdPR10-1 and MdPR10-2, respectively, to produce small interfering RNAs (siRNAs). The above vectors were transformed into GV3101 (an A. tumefaciens strain) by the heat shock transformation method. The cloning primers are listed in Supplemental Table 1.
Leaves from 4-week-old HF seedlings (resistant variety) and 4-week-old NGR196 seedlings (susceptible variety) were infiltrated by A. tumefaciens with a silencing or overexpression construct25. After agroinfiltration, the infiltrated seedlings were transferred to a fresh MS culture medium for 4 d to avoid wilting of the apple plantlets during Agrobacterium tumefaciens infiltration. After 4 days, the infiltrated apple plantlets were inoculated with a 2 × 105 CFU/mL pathogenic spore suspension, and the A. mali-inoculated seedlings were returned to fresh MS culture medium for 48 h to avoid wilting of the apple plantlets during fungal inoculation25.
Real-time PCR (RT-qPCR)
Total RNA of apple leaves was extracted with an EASY Spin Kit (Beijing Biomed Biotechnology Co., Ltd., China). The RNA was reverse-transcribed into cDNA using oligo-dT primers, which are listed in Supplemental Table 2. Real-time PCR analysis was performed using SuperReal PreMix Plus (SYBR Green) (Tiangen, FP205) under the following cycling procedure: 40 cycles of 95 °C for 10 s and 60 °C for 30 s (Applied Biosystems 7500). The relative RNA abundance was calculated using the 2−ΔΔCT method51, with MdActin as the reference gene. The specific primers for MdPR10-1, MdPR10-2, and the reference gene are listed in Supplemental Table 2.
Antifungal activity in vitro
The inhibition of A. mail mycelium growth by MdPR10-1 or MdPR10-2 in vitro was determined via incubation in Petri dishes by culturing A. mail spores on PDA medium containing purified GST protein, purified MdPR10-1 protein, or purified MdPR10-2 protein for 6 days. The temperature was controlled at 25 °C, and the cultures were kept in the dark.
Mycelium quantification was performed as described previously53. Photographs were taken with a scale bar after 6 days of growth on PDA, and the area of mycelium growth was calculated with ImageJ (http://imagej.nih.gov/ij/). The results for three independent biological replicates were obtained.
References
Zhang, C. X., Tian, Y. & Cong, P. H. Proteome analysis of pathogen-responsive proteins from apple leaves induced by the Alternaria Blotch Alternaria alternata. PLoS ONE. 10, e0122233 (2015).
Abe, K., Iwanami, H., Kotoda, N., Moriya, S. & Takahashi, S. Evaluation of apple genotypes and Malus species for resistance to Alternaria blotch caused by Alternaria alternata apple pathotype using detached-leaf method. Plant Breed. 129, 208–218 (2010).
Chung, K. R. Stress response and pathogenicity of the necrotrophic fungal pathogen Alternaria alternata. Scientifica. 2012, 635431 (2012).
Zhang, Q. et al. Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front Plant Sci. 8, 526 (2017).
Hugot, K., Aime, S., Conrod, S., Poupet, A. & Galiana, E. Developmental regulated mechanisms affect the ability of a fungal pathogen to infect and colonize tobacco leaves. Plant J. 20, 163–170 (1999).
Johnson, R. D. et al. Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM-toxin synthesis and pathogenicity. Mol. Plant Microbe 13, 742–753 (2020).
Moffett, P., Farnham, G., Peart, J. & Baulcombe, D. C. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519 (2020).
Jones, J. & Dangl, J. The plant immune system. Nature. 444, 323–329 (2006).
Choi, D. S., Hwang, I. S. & Hwang, B. K. Requirement of the cytosolic interaction between pathogenesis-related protein10 and leucine-rich repeat protein1 for cell death and defense signaling in pepper. Plant Cell. 24, 1675–1690 (2012).
Van Loon, L. C., Rep, M. & Pieterse, C. M. Significance of inducible defense-related proteins in infected plants. Annu Rev. Phytopathol. 44, 135–162 (2006).
Christensen, A. B. et al. The molecular characterization of two barley proteins establishes the novel PR-17 family of pathogenesis-related proteins. Mol. plant Pathol. 3, 135–144 (2002).
McGee, J. D., Hamer, J. E. & Hodges, T. K. Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Plant Microbe Interact. 14, 877–886 (2001).
Zhou, X. J., Lu, S., Xu, Y. H., Wang, J. W. & Chen, X. Y. A cotton cDNA (GaPR-10) encoding a pathogenesis-related 10 protein with in vitro ribonuclease activity. Plant Sci. 162, 629–636 (2002).
Hashimoto, M. et al. A novel rice PR10 protein, RSOsPR10, specififically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 45, 550–559 (2004).
Park, C. J. et al. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 37, 186–198 (2004).
Liu, J. J. & Ekramoddoullah, A. K. M. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 68, 3–13 (2006).
Breiteneder, H. et al. The gene coding for the major birch pollen allergen Betv1, is highly homologous to a pea disease resistance response gene. EMBO J. 8, 1935–1938 (1989).
Hashimoto, M. et al. The Role of alpha-synuclein assembly and metabolism in the pathogenesis of Lewy body disease. J. Mol. Neurosci. 24, 343–352 (2004).
Lo, S. C., Hipskind, J. D. & Nicholson, R. L. cDNA cloning of a sorghum pathogenesis-related protein (PR-10) and differential expression of defense-related genes following inoculation with Cochliobolus heterostrophus or Colletotrichum sublineolum. Mol. plant-microbe Interact. 12, 479–489 (1999).
Matton, D. P. & Brisson, N. Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol. plant-microbe Interact. 2, 325–331 (1989).
Pagliarani, G. et al. Genomic organisation of the Mal d 1 gene cluster on linkage group 16 in apple. Mol. Breed. 29, 759–778 (2012).
Somssich, I. E., Schmelzer, E., Bollmann, J. & Hahlbrock, K. Rapid activation by fungal elicitor of genes encoding “pathogenesis-related” proteins in cultured parsley cells. Proc. Natl Acad. Sci. USA. 83, 2427–2430 (1986).
Walter, M. H., Liu, J. W., Grand, C., Lamb, C. J. & Hess, D. Bean pathogenesis-related (PR) proteins deduced from elicitor-induced transcripts are members of a ubiquitous new class of conserved PR proteins including pollen allergens. Mol. Gen. Genet. 222, 353–360 (1990).
Warner, S. A., Scott, R. & Draper, J. Characterisation of a wound-induced transcript from the monocot asparagus that shares similarity with a class of intracellular pathogenesis-related (PR) proteins. Plant Mol. Biol. 19, 555–561 (1992).
Jung, Y. H. et al. The rice (Oryza sativa) blast lesion mimic mutant, blm, may confer resistance to blast pathogens by triggering multiple defense-associated signaling pathways. Plant Physiol. Biochem. 43, 397–406 (2005).
Jung, Y. H. et al. Differential expression of defense/stressrelated marker proteins in leaves of a unique rice blast lesion mimic mutant (blm). J. Proteome Res. 5, 2586–2598 (2005).
Besbes, F., Habegger, R. and Schwab, W. Induction of PR-10 genes an meta- bolites in strawberry plants in response to Verticillium dahliae infection. BMC Plant Biol. 19, 128 (2019).
Somssich, I. E., Schmelzer, E., Kawalleck, P. & Hahlbrock, K. Gene structure and in situ transcript localization of pathogenesis-related protein 1 in parsley. Mol. Gen. Genet 213, 93–98, https://doi.org/10.1007/BF00333403 (1988).
Breiteneder, H. et al. Molecular characterization of Api g 1, the major allergen of celery (Apium graveolens), and its immunological and structural relationships to a group of 17-kDa tree pollen allergens. Eur. J. Biochem 233, 484–489, https://doi.org/10.1111/j.1432-1033.1995.484_2.x (2010).
Vanek-Krebitz, M. et al. Cloning and sequencing of Mal d 1, the major allergen from apple (Malus domestica), and its immunological relationship to Bet v 1, the major birch pollen allergen. Biochem Biophys. Res Commun. 214, 538–551, https://doi.org/10.1006/bbrc.1995.2320 (1995).
Puhringer, H. et al. The promoter of an apple Ypr10 gene, encoding the major allergen Mal d 1, is stress- and pathogen-inducible. Plant Sci. 152, 35–50 (2000).
Jung, H. W. & Hwang, B. K. The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol. Plant Pathol. 8, 503–514 (2007).
Choi, H. W., Kim, Y. J. & Hwang, B. K. The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant Microbe Interact. 24, 68–78 (2011).
Fernandes, H., Michalska, K., Sikorski, M. and Jaskolski, M. Structural and functional aspects of PR-10 proteins. FEBS J. 280, 1169–1199 (2013).
Fujimoto, Y. et al. Purifification and cDNA cloning of cytokinin-specifific binding protein from mung bean (Vigna radiata). Eur. J. Biochem. 258, 794–802 (1998).
Mogensen, J. E., Wimmer, R., Larsen, J. N., Spangfort, M. D. & Otzen, D. E. The major birch allergen, Bet v 1, shows affifinity for a broad spectrum of physiological ligands. J. Biol. Chem. 277, 23684–23692 (2002).
Markovic´-Housley, Z. et al. Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J. Mol. Biol. 325, 123–133 (2003).
Koistinen, K. M. et al. Birch PR-10c interacts with several biologically important ligands. Phytochemistry 66, 2524–2533 (2005).
Harteveld, D., Akinsanmi, O., Chandra, K. & Drenth, A. Timing of infection and development of alternaria diseases in the canopy of apple trees. Plant Dis. 98, 401–408 (2014).
Sawamura, K. Studies on apple Alternaria blotch caused by Alternaria mali Roberts. Bull. Fac. Agric. Hirosaki Univ. 18, 152–235 (1972).
Li, Y. et al. Interactions of apple and the Alternaria alternata apple pathotype. Crit. Rev. Plant Sci. 32, 141–150 (2013).
Hwang, B., Yun, J. & Kim, Z. Geographic variation of esterase isozymes in populations of Alternaria mali on apple leaves. J. Phytopathol. 119, 225–231 (1987).
Filajdic, N. & Sutton, T. Identification and distribution of Alternaria mali on apples in North Carolina and susceptibility of different varieties of apples to Alternaria blotch. Plant Dis. 75, 1045–1048 (1991).
Gagkaeva, T. & Levitin, M. Identification of the pathogen causing leaf spots on apple trees in Krasnodar province orchards. Mikol. Fitopatol. 34, 58–62 (2000).
Bulajic, A., Filajdic, N., Babovic, M. & Sutton, T. First report of Alternaria mali on apples in Yugoslavia. Plant Dis. 80, 709 (1996).
Soleimani, M. & Esmailzadeh, M. First report of Alternaria mali causing apple leaf blotch disease in Iran. Australas. Plant Dis. Notes. 2, 57–58 (2007).
Ozgonen, H. & Karaca, G. First report of Alternaria mali causing necrotic leaf spot of apples in Turkey. Plant Pathol. 55, 578 (2006).
Rollemberg, C., Fayad, F., Hamada, N. & De Mio, L. First report of Alternaria mali on apples in Brazil. Phytopathology 101, 155 (2011).
Wang, M., Weiberg, A., Dellota, E., Yamane, D. & Jin, H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 14, 421–428 (2017).
Gachon, C. & Saindrenan, P. Real-time PCR monitoring of fungal development in Arabidopsis thaliana infected by Alternaria brassicicola and Botrytis cinerea. Plant Physiol. Biochem. 42, 367–371 (2004).
Livak, K. & Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001).
Bai, S., Kasai, A., Yamada, K., Li, T. & Harada, T. A mobile signal transported over a long distance induces systemic transcriptional gene silencing in a grafted partner. J. Exp. Bot. 62, 4561–4570 (2011).
Song, N., et al. An erf2-like transcription factor regulates production of the defense sesquiterpene capsidiol upon alternaria alternata infection. J. Exp Bot. 20, 5895–5908 (2019).
Acknowledgements
We thank Professor LiYun Guo from China Agricultural University for providing us with the apple leaf spot fungal strain Alternaria alternata f. sp. mali ALT1, and we thank Professor XiaoQiong Zhu, also from China Agricultural University, for providing the strains A. alternata f. sp. mali GBYB2, BXSB5, and BXSB7.
Author information
Authors and Affiliations
Contributions
Q.L.Z. and T.L. conceived the research plans; Q.L.Z. performed most of the experiments; H.Y.W., C.R.X., and W.Q.F. performed the rest of the experiments; Q.L.Z. analyzed the data; and Q.L.Z. and T.L. wrote the article. This work was funded by the National Natural Science Foundation of China (32002013) and supported by the Chinese Universities Scientific Fund (2021TC074) and The 2115 Talent Development Program of China Agricultural University.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Q., Xu, C., Wei, H. et al. Two pathogenesis-related proteins interact with leucine-rich repeat proteins to promote Alternaria leaf spot resistance in apple. Hortic Res 8, 219 (2021). https://doi.org/10.1038/s41438-021-00654-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00654-4